I recently read through a lecture slide deck called ‘Judging the Evidence’ by Adrian Sleigh for a course PUBH7001 Introduction to Epidemiology, April 30, 2001.
It had a lot of great material in it but I especially liked the section ‘CRITIQUE OF AN EPIDEMIOLOGIC STUDY’ and slide 11 ‘Quantity of data, duplication’ which says:
Set clear criteria for admission of studies to your ‘judgement of evidence’
Devise ways to tabulate the information
‘Evidence tables’ show key features of design
(source, sample and study pop, N)
exposures-outcomes measured
observation methods
confounding
key results
I thought this was a great idea, to build a database for keeping ‘evidence tables’ for each study I read.
I then read through all the slides. There is a lot of great information here, but it was spread out across the narrative. I realised I wanted to collate these into a ‘evidence table’. I also compared this with my understanding of the Ecological Metadata Language (EML) schema and the ‘ANU Data Analysis Plan Template’ and have put together a bit of a ‘cross-walk’ that lets me combine all this info and create a evidence table (database).
I have started to use the database I built which uses EML concepts heavily and I include some these other ideas into my free data_inventory application for a web2py database https://github.com/ivanhanigan/data_inventory.
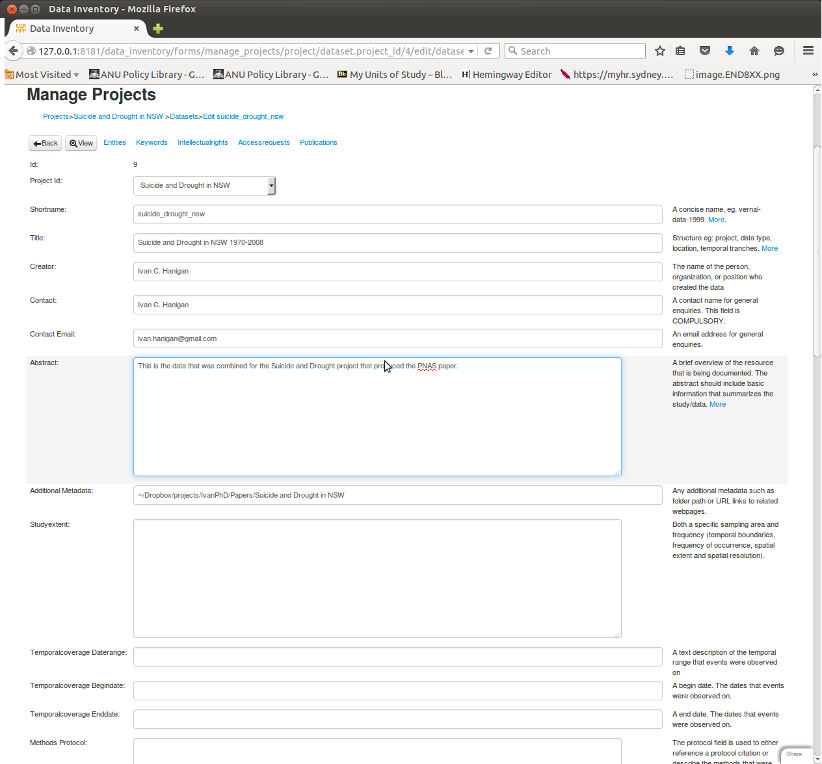
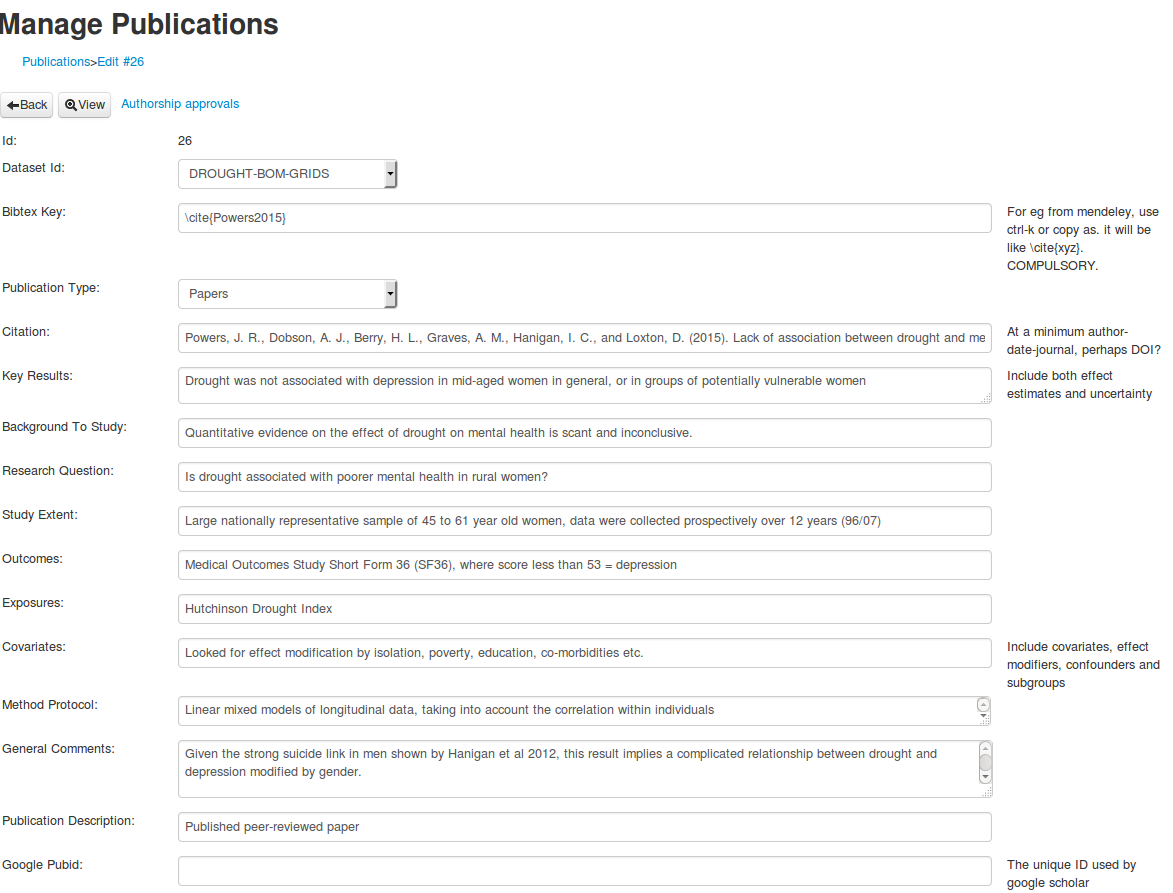
It is a webform style data entry interface, and I think good for these ‘evidence tables’.
In the first instance I piggy back a lot of the elements into single EML tags, especially the abstract.
This may make it hard to parse. The simple solution is to try to keep each element on a seperate line of the absract.
The key info for an evidence table entry per study
| EML | ANU | Adrian_Sleigh |
| dataset/title | Study name | |
| dataset/creator | Person conducting analysis | |
| project/personnel/[data_owner or orginator] | Chief investigator | |
| dataset/abstract | Background to the study | Purpose of Study |
| | Study research question | |
| | Specific hypothesis under study | |
| | outcomes of interest/ Exposure variables /Covariates | exposures-outcomes measured |
| | | key results |
| dataset/studyextent | Study population | Study Setting |
| | | source / sample and study pop |
| dataset/temporalcoverage | Duration of study | |
| dataset/methods_protocol | Study Type | Type of study |
| dataset/sampling_desc | | Subject Selection |
| dataset/methods_steps | analytical strategy | Statistical procedures |
| | exposures/ potential confounders or effect modifiers | Confounding |
| entity/numberOfRecords | Number study subjects | N |
| dataset/distribution_methods | dissemination strategy | |
Here is a screen shot of my data inventory data entry form