- Yesterday I met with Sarunya Sujaritpong a PhD student working with spatially structured time-series models as described previously
- Her supervisor Keith Dear has given me a lot of good stats advice in the past and one bit I keep thinking about is that a time series model can be fit for multiple spatial areal units of the same city and residual spatial auto-correlation in the errors should not be too much of a concern
- The problem would be if you get strong spatial autocorrelation of the residuals this indicates that the assumption of independent errors is violated and you will have tighter confidence intervals around the coefficients of interest than is really the case, inflating the signficance estimated for the relative risk
- The beta coefficient itself shouldn’t be affected.
- So long as biostatisticians like Keith are comfortable with not addressing this issue in spatially structured time-series that is great but I see people are starting to include this in their models
- To date I’ve mostly seen it done in spatial (cross sectional) data analysis, not spatial times-series
- I’m preparing for the day when I might need to address this for a reviewer and would like to know what to do about it in case that happens
- So I asked Sarunya for a discussion about her research
Sarunya’s model is essentially like this
R Code:
fit <- glm(deaths ~ pollutant1 + pollutant2 + pollutant ... +
ns(temp, df) + ns(humidity, df) +
ns(time, df = 7*numYears) +
SLA * ns(time, df = 2),
data = analyte, family = poisson
)
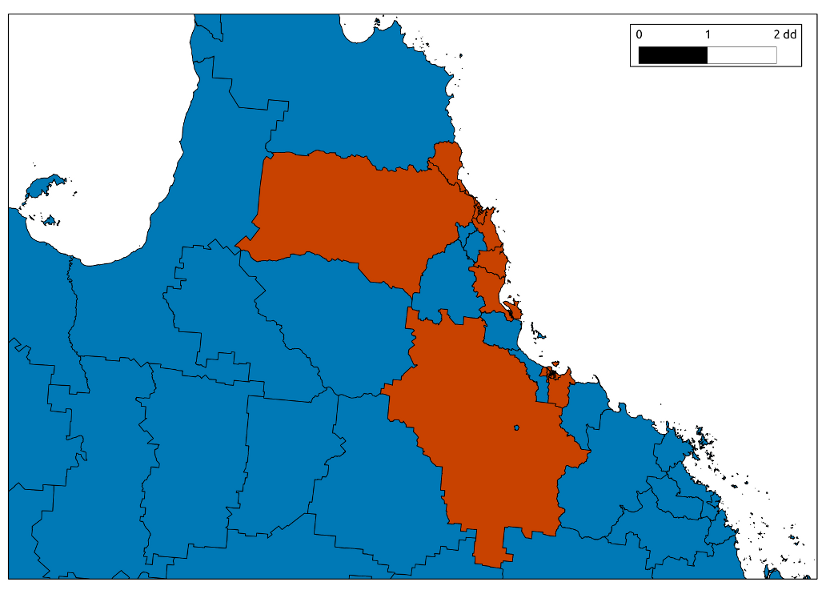
- SLA is Statistical Local Area (now called SA2, like a suburb)
- Sarunya explained that the research question was if the magnitude of the coeff on pollutant1 differed between this model and the old style of model where an entire city is used as the unit of analysis per day and exposure estimates are calculated as averages across several monitoring stations in the city
- turns out that this comparison is still valid EVEN IF THE STANDARD ERROR IS BIASED DUE TO RESIDUAL SPATIAL AUTOCORRELATION
- therefore this study avoids the issue by it’s intentional design to compare betas not se
- Interestingly Sarunya explained that the stats theory suggests that even if the exposure precision is increased (exposure misclassification bias is decreased) the se on the beta will not be affected.
- this is fascinating in itself, but a separate issue for another post
Conclusions
- So it looks like the extent a study might need to consider the issue of potential residual spatial autocorrelation depends alot on what questions are being asked and what inferences will be attempted from the results
- if the aim of the study is to estimate the magnitude AND CONFIDENCE INTERVALS of an exposure’s relative risk (especially some novel exposure such as interstellar space dust across the suburbs) then the issue might become important to address.
THANKS Sarunya!