We recently hit an issue when using morpho to enter metadata for a large number of variables (~200). The GUI form for entering definitions and units steps through each variable, but at about 60 or 70 it starts to slow down. By the time we got to 160 it was taking more than 5 minutes to change to the next variable. To safegaurd against losing work, we kept hitting “save for later” but this got slower and apeared to freeze at the last minute… Not sure if that last save worked at all.
So I;ve come back once more to the ROpenSci EML package which is looking like a really useful way to build metadata elements automatically, with Morpho being used to proved augmentation and finesse the documents.
First thing I tried was the constructed Column Definitions and Unit Definitions example from the README
R Code:
#require(devtools)
#install_github("EML", "ROpenSci")
require(EML)
# The example from orig doco
dat = data.set(river = c("SAC", "SAC", "AM"),
spp = c("king", "king", "ccho"),
stg = c("smolt", "parr", "smolt"),
ct = c(293L, 410L, 210L),
col.defs = c("River site used for collection",
"Species common name",
"Life Stage",
"count of live fish in traps"),
unit.defs = list(c(SAC = "The Sacramento River",
AM = "The American River"),
c(king = "King Salmon",
ccho = "Coho Salmon"),
c(parr = "third life stage",
smolt = "fourth life stage"),
"number"))
str(dat)
eml_config(creator="Carl Boettiger <cboettig@gmail.com>")
eml_write(dat, file = "inst/doc/EML_example.xml")
# now you can import this to morpho and have a look
# note that for morpho 1.8 it wants to change the EML version from 2.1.1 to 2.1.0
# and it can't show the data yet
# so what we need to do is write this as a file to the morpho database
# save and close, note which number it assigned
dat <- data.frame(dat)
morpho_db <- "~/.morpho/profiles/hanigan/data/hanigan/"
maxid <- 1+max(as.numeric(dir(morpho_db)))
filename <- file.path(morpho_db,maxid)
# what is the number?
filename
write.csv(dat, filename, row.names =F, quote=F)
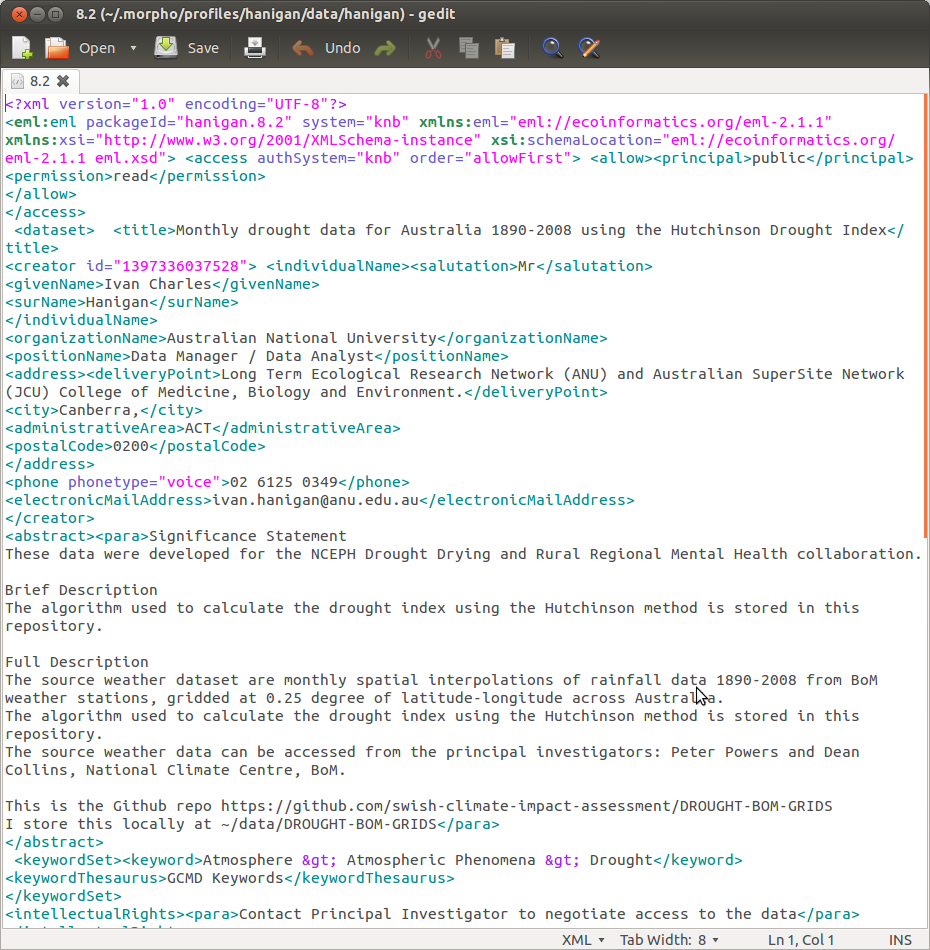
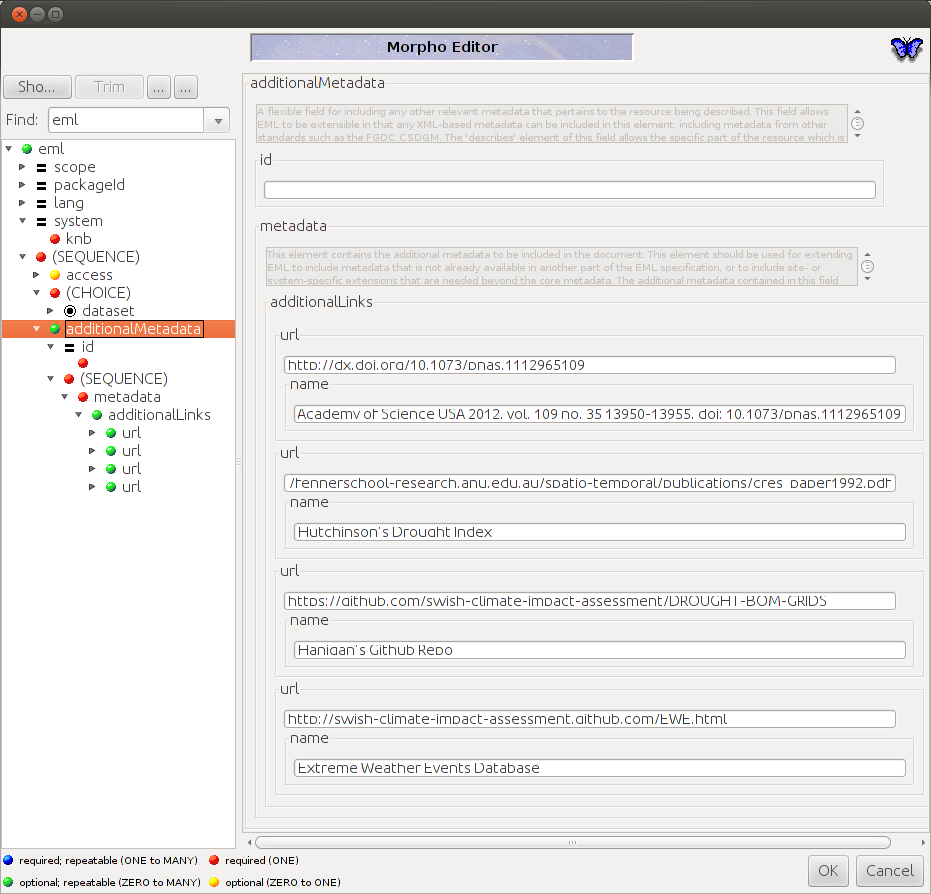
So now to finish what we need to add into the EML that morpho has created (22.1 in my case) just needs the reference to the dataTable.
EML Code:
...
</dataFormat>
<distribution scope="document"> <online> <url function="download">ecogrid://knb/hanigan.22.1</url>
</online>
<access authSystem="knb" order="denyFirst"><allow><principal>uid=datalibrarian,o=unaffiliated,dc=ecoinformatics,dc=org</principal>
<permission>all</permission>
</allow>
<allow><principal>uid=hanigan,o=unaffiliated,dc=ecoinformatics,dc=org</principal>
<permission>read</permission>
</allow>
</access>
</distribution>
</physical>
...
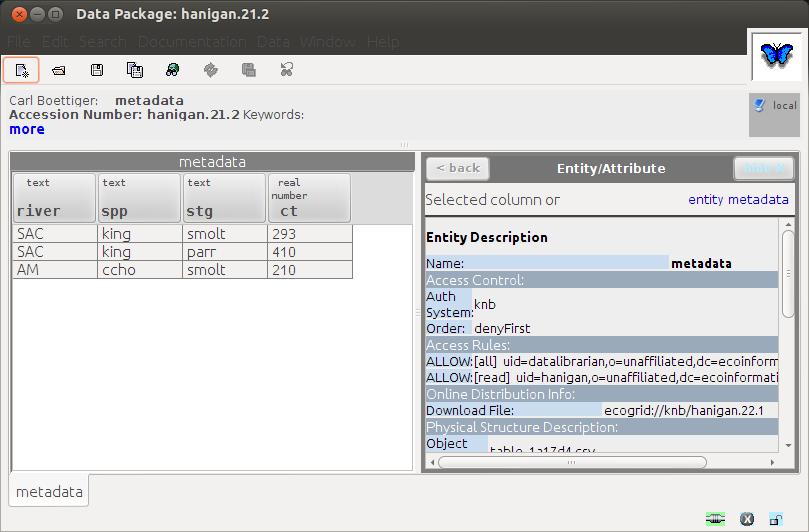
Which seems to have worked when we open it up again:
So now let;s try a large nuber of variables:
R Code:
# add lots of cols
for(i in 5:100){
dat[,i] <- sample(rnorm(100,1,2), 3)
}
str(dat)
## $ V95 : num 1.5708 -0.0936 2.2324
## $ V96 : num 1.79 5.4 1.62
## $ V97 : num -1.141 0.653 5.365
## $ V98 : num 1.738 -1.046 -0.135
## $ V99 : num 3.6 -0.738 -1.877
## [list output truncated]
# firstly I make a liset of the unit definitions for the example
unit.defs <- list(c(SAC = "The Sacramento River",
AM = "The American River"),
c(king = "King Salmon",
ccho = "Coho Salmon"),
c(parr = "third life stage",
smolt = "fourth life stage"))
# then I add to it the definition for the constructed variables
unit.defs[[3]]
for(i in 4:100){
unit.defs[[i]] <- "number"
}
unit.defs
# and this can be passed to the data.set constructor
dat <- data.set(dat,
col.defs = c("River site used for collection",
"Species common name",
"Life Stage",
"count of live fish in traps",
c(rep("count stuff", 95))),
unit.defs = unit.defs
)
str(dat)
eml_config(creator="Ivan Charles Hanigan <ivan.hanigan@gmail.com>")
eml_write(dat, file = "inst/doc/EML_example_wide.xml")
# import to morpho, save and close
# create the dataset for morphos database
dat <- data.frame(dat)
morpho_db <- "~/.morpho/profiles/hanigan/data/hanigan/"
maxid <- 1+max(as.numeric(dir(morpho_db)))
filename <- file.path(morpho_db,maxid)
# what is the number?
filename
write.csv(dat, filename, row.names =F, quote=F)
# now add this into the EML morpho has created (25.2 in my case)
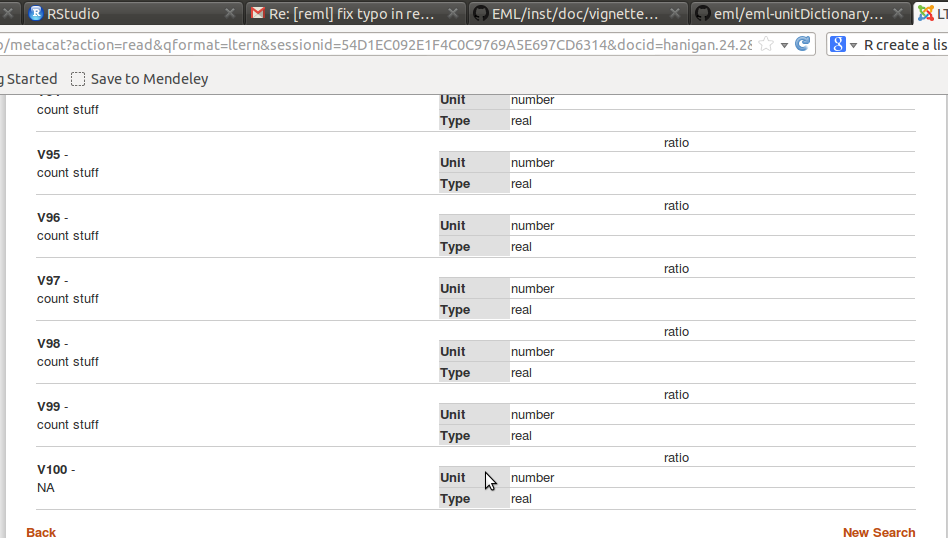
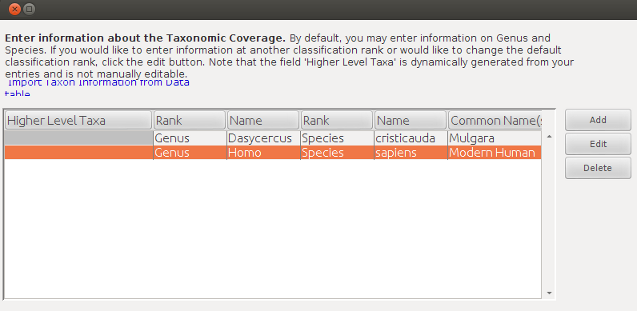
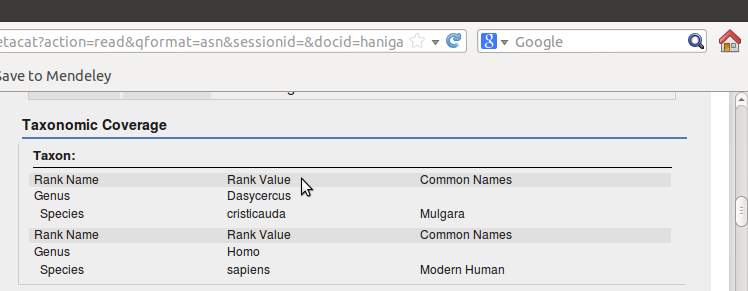
Which now seems to have attached the variable defintions and dataTable adequately.
name: using-reml-to-input-large-number-of-column-descriptions layout: post title: Using Reml To Input Large Number Of Column Descriptions date: 2014-04-24 categories:
-
Data Documentation
We recently hit an issue when using morpho to enter metadata for a large number of variables (~200). The GUI form for entering definitions and units steps through each variable, but at about 60 or 70 it starts to slow down. By the time we got to 160 it was taking more than 5 minutes to change to the next variable. To safegaurd against losing work, we kept hitting “save for later” but this got slower and apeared to freeze at the last minute… Not sure if that last save worked at all.
So I;ve come back once more to the ROpenSci EML package which is looking like a really useful way to build metadata elements automatically, with Morpho being used to proved augmentation and finesse the documents.
First thing I tried was the constructed Column Definitions and Unit Definitions example from the README
R Code:
#require(devtools)
#install_github("EML", "ROpenSci")
require(EML)
# The example from orig doco
dat = data.set(river = c("SAC", "SAC", "AM"),
spp = c("king", "king", "ccho"),
stg = c("smolt", "parr", "smolt"),
ct = c(293L, 410L, 210L),
col.defs = c("River site used for collection",
"Species common name",
"Life Stage",
"count of live fish in traps"),
unit.defs = list(c(SAC = "The Sacramento River",
AM = "The American River"),
c(king = "King Salmon",
ccho = "Coho Salmon"),
c(parr = "third life stage",
smolt = "fourth life stage"),
"number"))
str(dat)
eml_config(creator="Carl Boettiger <cboettig@gmail.com>")
eml_write(dat, file = "inst/doc/EML_example.xml")
# now you can import this to morpho and have a look
# note that for morpho 1.8 it wants to change the EML version from 2.1.1 to 2.1.0
# and it can't show the data yet
# so what we need to do is write this as a file to the morpho database
# save and close, note which number it assigned
dat <- data.frame(dat)
morpho_db <- "~/.morpho/profiles/hanigan/data/hanigan/"
maxid <- 1+max(as.numeric(dir(morpho_db)))
filename <- file.path(morpho_db,maxid)
# what is the number?
filename
write.csv(dat, filename, row.names =F, quote=F)
So now to finish what we need to add into the EML that morpho has created (22.1 in my case) just needs the reference to the dataTable.
EML Code:
...
</dataFormat>
<distribution scope="document"> <online> <url function="download">ecogrid://knb/hanigan.22.1</url>
</online>
<access authSystem="knb" order="denyFirst"><allow><principal>uid=datalibrarian,o=unaffiliated,dc=ecoinformatics,dc=org</principal>
<permission>all</permission>
</allow>
<allow><principal>uid=hanigan,o=unaffiliated,dc=ecoinformatics,dc=org</principal>
<permission>read</permission>
</allow>
</access>
</distribution>
</physical>
...
Which seems to have worked when we open it up again:

So now let;s try a large nuber of variables:
R Code:
# add lots of cols
for(i in 5:100){
dat[,i] <- sample(rnorm(100,1,2), 3)
}
str(dat)
## $ V95 : num 1.5708 -0.0936 2.2324
## $ V96 : num 1.79 5.4 1.62
## $ V97 : num -1.141 0.653 5.365
## $ V98 : num 1.738 -1.046 -0.135
## $ V99 : num 3.6 -0.738 -1.877
## [list output truncated]
unit.defs = list(c(SAC = "The Sacramento River",
AM = "The American River"),
c(king = "King Salmon",
ccho = "Coho Salmon"),
c(parr = "third life stage",
smolt = "fourth life stage"))
unit.defs[[3]]
for(i in 4:100){
unit.defs[[i]] <- "number"
}
unit.defs
dat = data.set(dat,
col.defs = c("River site used for collection",
"Species common name",
"Life Stage",
"count of live fish in traps",
c(rep("count stuff", 95))
),
unit.defs = unit.defs
)
str(dat)
eml_config(creator="Ivan Charles Hanigan <ivan.hanigan@gmail.com>")
eml_write(dat, file = "inst/doc/EML_example_wide.xml")
# import to morpho, save and close
dat <- data.frame(dat)
morpho_db <- "~/.morpho/profiles/hanigan/data/hanigan/"
maxid <- 1+max(as.numeric(dir(morpho_db)))
filename <- file.path(morpho_db,maxid)
# what is the number?
filename
write.csv(dat, filename, row.names =F, quote=F)
# now add this into the EML morpho has created (25.2 in my case)
Which now seems to have attached the variable defintions and dataTable adequately.