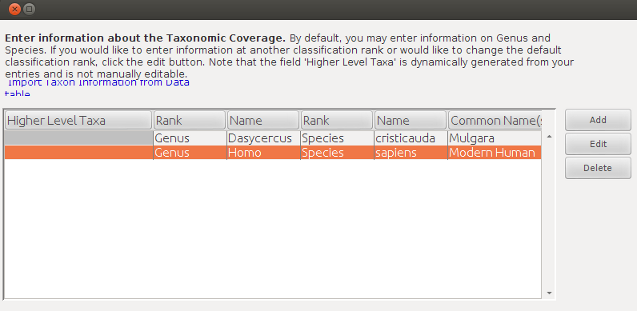
Morpho is a pretty minimal editor for EML really. It gives you a set of generically useful data entry forms but sometimes a specific task is better achieved through edits made directly to the XML document. An example of this is inserting a large number of species names to the taxonomic coverage module. The form to include these requires individual data entry for each species.
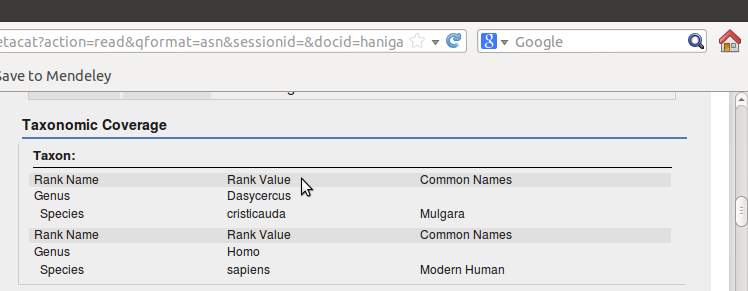
Which looks like this when published
Go to the morpho catalogue (found at ~/.morpho/profiles/hanigan/data/hanigan/) and take a look at how the XML is constructed.
XML Code:
<taxonomicCoverage>
<taxonomicClassification>
<taxonRankName>Genus</taxonRankName>
<taxonRankValue>Dasycercus</taxonRankValue>
<taxonomicClassification>
<taxonRankName>Species</taxonRankName>
<taxonRankValue>cristicauda</taxonRankValue>
<commonName>Mulgara</commonName>
</taxonomicClassification>
</taxonomicClassification>
<taxonomicClassification>
<taxonRankName>Genus</taxonRankName>
<taxonRankValue>Homo</taxonRankValue>
<taxonomicClassification>
<taxonRankName>Species</taxonRankName>
<taxonRankValue>sapiens</taxonRankValue>
<commonName>Modern Human</commonName>
</taxonomicClassification>
</taxonomicClassification>
</taxonomicCoverage>
SO how to insert a large number of these
One could use the taxon import feature to import the details from a file if there is a large list. Morpho’s taxon import feature does not correctly import more than one column of taxon data so if you have Genus and Species to enter you will actually need to combine Genus and Species into a binomial Species rank in one column (beware that if the data are sourced from a datafile that uses the underscore to seperate the words then these will not be correctly imported).
Once you have formated your input list, then import this as a new data table and go to the taxonomic coverage form under the documentation menu. Click on the option to “import taxon information from data table” and select the appropriate column, selecting ‘species’ for the class. This will populate the taxonomicCoverage module in the EML. You can now remove that data table from the package to be tidy.
This is what it looks like if you combine genus and species in the single column.
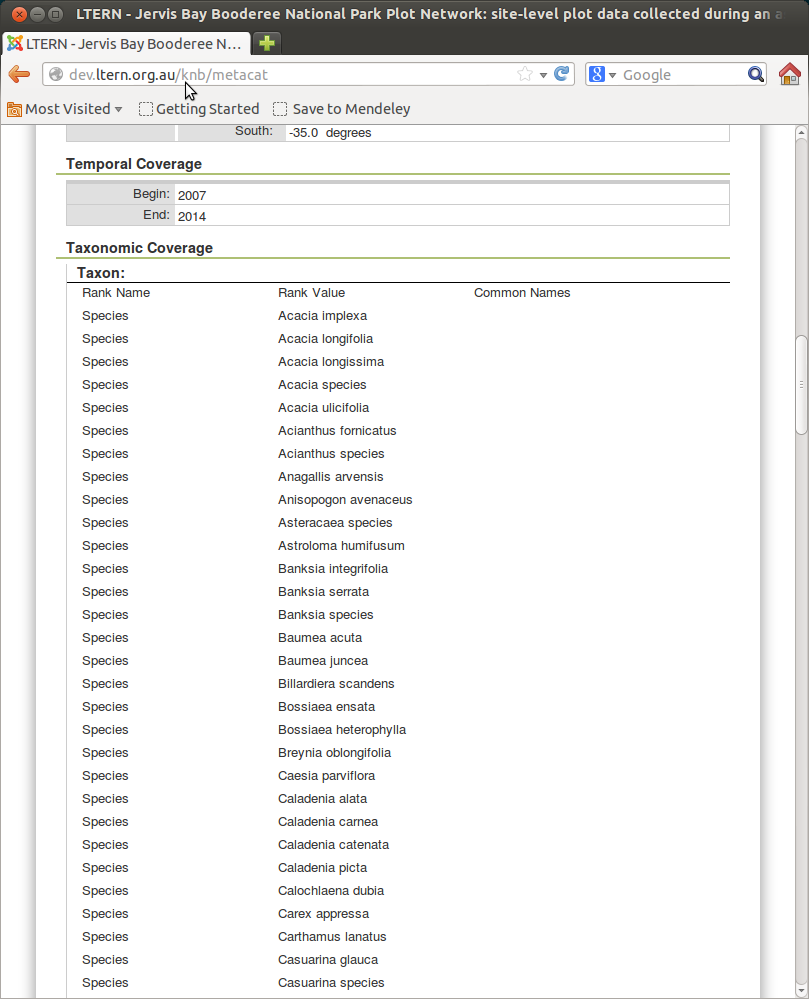
And here is the XML
XML Code:
<taxonomicCoverage scope="document">
<taxonomicClassification>
<taxonRankName>Species</taxonRankName>
<taxonRankValue>Abelmoschus moschatus</taxonRankValue>
</taxonomicClassification>
<taxonomicClassification>
<taxonRankName>Species</taxonRankName>
<taxonRankValue>Abrus pector</taxonRankValue>
</taxonomicClassification>
<taxonomicClassification>
<taxonRankName>Species</taxonRankName>
<taxonRankValue>Abrus precatorius</taxonRankValue>
</taxonomicClassification>
Morpho has problems subsequently editing a very long list
We found that if a very large amount of taxonomic information is entered into Morpho we had issues modifying it. When you click on Documentation > Taxonomic Coverage to try and go in and edit nothing will happen. Morpho crashes when trying to open the Taxonomic Coverage because the list is long enough to cause “Out of Memory” error with the default configuration of Java heap space. It is a Morpho bug. The workaround is to edit the XML file manually.





