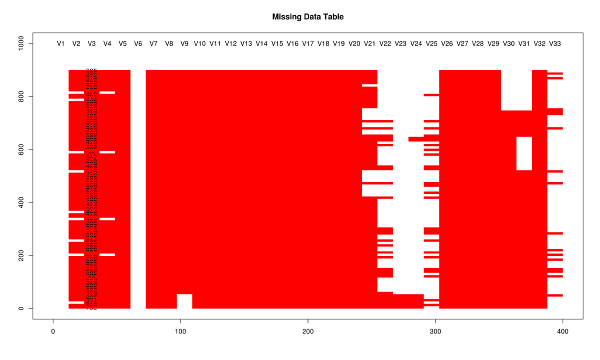
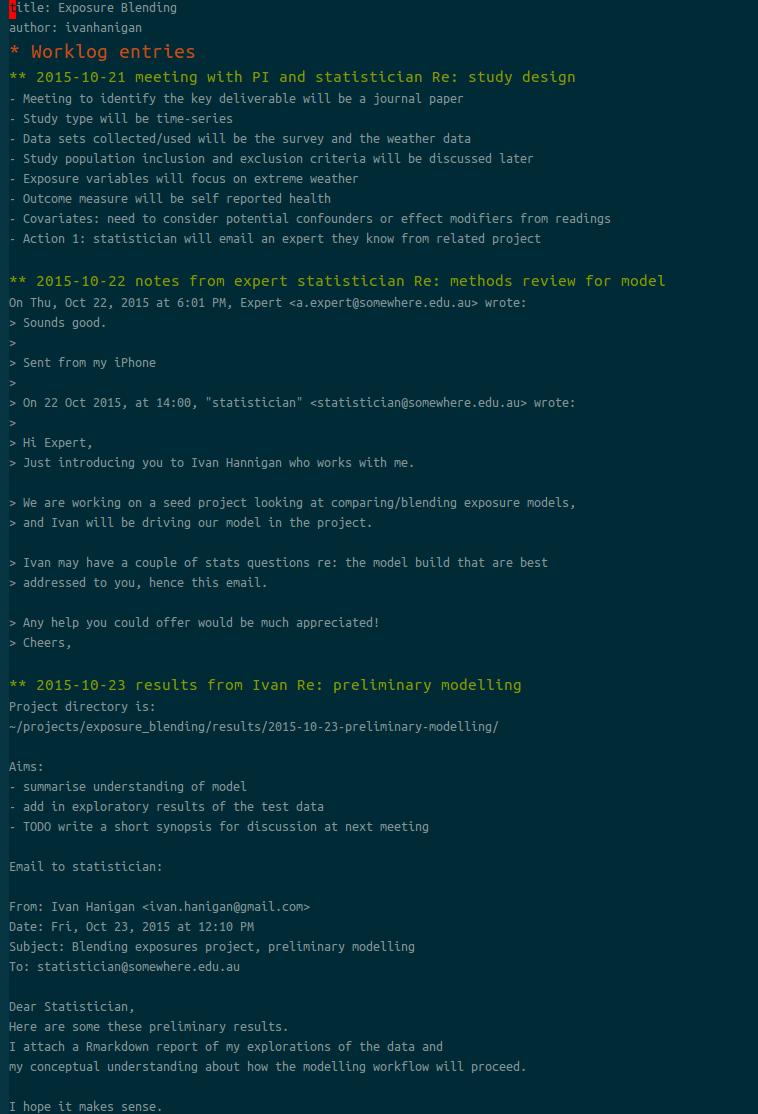
This post focuses on decision making during statistical modelling. The case of investigating effect modification is used as an example. The analyst has several options available to them when constructing the model parametrisation for adjustment to explain modification effectively. Unlike confounding (in which the criterion of substantial magnitude change of the effect estimate when controlling for a third variable can be easily assessed), models including effect modification can be hard to interpret. Taking account of effect modification becomes increasingly important when modelling complex interactions.
If an effect moderator is present then the relationship between exposure and response varies between different levels of the moderator. An example is provided by a paper we published, in which the relationship between proximity to wetlands and Ross River virus infection was found to be different for towns and rural areas, where the ‘urban’ variable is the ‘effect moderator’.
There are three common ways for data analysts to address this question in statistical models:
- multiple models for each group,
- interaction terms or
- re-parametrisation to explicitly depict interactions.
The first way is to split apart the dataset and conduct separate analyses of multiple groups. For example, one could run the regression in the urban zone and the rural zone seperately and see if there were different exposure response functions in the two models. This was the approach of our paper. This approach has the strength that it is simple to do and yeilds results that are easy to interpret. A limitation of this method is that by splitting the dataset one loses degrees of freedom and therefore statistical power.
The second approach (which removes that limitation of the first option) is to fit interaction terms. The example shown in Figure 1 is a multiplicative interaction model (Brambor 2006). This approach was not taken in the models reported in our paper, but can easily be implemented and shows that the function of distance was estimated to have different ‘slopes’ in each of the dichotomous urban groups.
The statistical method can be easily implemented in software by including a multiplicative term between two variables, however in practice the resulting post estimation regression outputs can be difficult to interpret. For example, say one wants to calculate the effect and standard error for exposure X on health outcome Y with the interaction of the effect modifier Z. The form of this model can be written as:
\[Y ~ \beta_{1}X + \beta_{2}Z + \beta_{3}XZ\]where B1, B2 and B3 are the regression coefficients estimated and the term XZ is the interaction between exposure and effect modifier.
The difficulty for interpretation comes when using this method for calculating the marginal effect of X on Y and the conditional standard error. The specific method described in Brambor et al. is:
- Calculate the coefficients and the variance-covariance matrix from the regression model
- The marginal effect is $\beta_{1} + \beta_{3}XZ$ where Z is the level of the modifying factor (0 or 1 in the dichotomous effect modifier case)
- The conditional standard error is:
Therefore the strengths of this approach is that it does not reduce degrees of freedom and is straightforward to specify the model in standard statistical software packages. The limitations are related to interpretation of the resulting coefficients for both the main effects and the marginal effects, and standard errors for these.
The third approach available to analysts makes it easier to access the resulting regression output. This method was employed in Paper 1 in the final modelling phase in which estimates were calculated for the different drought exposure-response funcitons in each of the sub-groups. In the terms of Figure \ref{fig:effectmod.png} it is simple to:
- Calculate X1 = X * Z (i.e = exposure for condition is met, zero otherwise)
- Calculate X0 = X * (1-Z) (i.e. = exposure for NON-condition, zero for condition is TRUE)
- Instead of X, Z and XZ, fit X1, X0 and Z. This model also contains three parameters and captures the same interactions as it is the same model with a different parametrisation. The standard errors for the X1 and X0 are calculated directly from the regression.
This method is much easier to implement and interpret. This is also considerably more flexible than the other two approaches. A limitation remains for this method in that the pre-processing steps required are more complicated, and there are inherently more possibilities for the data analyst to make errors in writing their code as they make these changes to the analytical data.
# Demonstrating this with the data from the paper (available in table 2)
## the following code shows the different parametrisations for the effect modification by urban
## we show that the coeffs and se are equivalent but that the psuedo-R
## squared will be better when including all our data in stratified analysis
# model 0 effect in eastern
d_eastern
fit <- glm(cases~ buffer + offset(log(1+pops)),family='poisson', data=d_eastern )
summa <- summary(fit)
summa
## Coefficients:
## Estimate Std. Error z value Pr(>|z|)
## (Intercept) -4.82425 0.14451 -33.382 < 2e-16 ***
## buffer -0.24702 0.07921 -3.119 0.00182 **
# model 1 effect in urban
fit1 <- glm(cases~ buffer + offset(log(1+pops)),family='poisson', data=d_urban )
summa <- summary(fit1)
summa
## Coefficients:
## Estimate Std. Error z value Pr(>|z|)
## (Intercept) -5.52363 0.33354 -16.561 <2e-16 ***
## buffer 0.03853 0.06352 0.607 0.544
# step 1, combine the urban and rural data
d_eastern$urban <- 0
d_urban$urban <- 1
dat2 <- rbind(d_eastern, d_urban)
str(dat2)
dat2
# model 2 is a multiplicative term
fit2 <- glm(cases ~ buffer * urban + offset(log(1+pops)), family = 'poisson', data = dat2)
summa <- summary(fit2)
summa
## Coefficients:
## Estimate Std. Error z value Pr(>|z|)
## (Intercept) -4.82425 0.14451 -33.382 < 2e-16 ***
## buffer -0.24702 0.07921 -3.119 0.00182 **
## urban -0.69938 0.36350 -1.924 0.05435 .
## buffer:urban 0.28555 0.10153 2.812 0.00492 **
# the coeff on buffer is for urban = 0 is main effect
# the coeff on buffer:urban is for urban = 1 is the marginal effect
b1 <- summa$coeff[2,1]
b3 <- summa$coeff[4,1]
b1 + b3
# 0.0385268
# but what about that p-value? and the se?
str(fit2)
fit2_vcov <- vcov(fit2)
fit2_vcov
# now calculate the conditional standard error for the marginal effect of buffer for the value of the modifying variable (Z, urban =1)
varb1<-fit2_vcov[2,2]
varb3<-fit2_vcov[4,4]
covarb1b3<-fit2_vcov[2,4]
Z<-1
conditional_se <- sqrt(varb1+varb3*(Z^2)+2*Z*covarb1b3)
conditional_se
# model 3 is the re-parametrisation
dat2$buffer_urban <- dat2$buffer * dat2$urban
dat2$buffer_eastern <- dat2$buffer * (1-dat2$urban)
fit3 <- glm(cases ~ buffer_urban + buffer_eastern + urban + offset(log(1+pops)), family = 'poisson', data = dat2)
summa <- summary(fit3)
summa
## Coefficients:
## Estimate Std. Error z value Pr(>|z|)
## (Intercept) -4.82425 0.14451 -33.382 < 2e-16 ***
## buffer_urban 0.03853 0.06352 0.607 0.54416
## buffer_eastern -0.24702 0.07921 -3.119 0.00182 **
## urban -0.69938 0.36350 -1.924 0.05435 .