Disentangle Things
Table of Contents
- 1 morpho-and-reml-boilerplate-streamline-the-process-of-metadata-entry
- 1.1 Background
- 1.2 Speed and Rigour
- 1.3 Analysts can often trade-off completeness of documentation for speed
- 1.4 Librarians produce gold plated documentation and can take longer to produce this
- 1.5 An example
- 1.6 Embracing Inaccuracy and Incompleteness
- 1.7 Aim
- 1.8 Step 1: load a simple example dataset
- 1.9 Step 2 create a function to deliver the minimal metadata object
- 1.10 remlboilerplate-code
- 1.11 remlboilerplate-test-code
- 1.12 Results: This loads into Morpho with some errors
- 1.13 Conclusions
1 morpho-and-reml-boilerplate-streamline-the-process-of-metadata-entry
1.1 Background
- The Morpho/Metacat system is great for a data repository
- Morpho also claims to be suitable for Ecologists to document their data
- But in my experience it leaves a little to be desired in ease of use for both purposes
- Specifically the speed that documentation can be entered into Morpho is slow
- This post is a first attempt to create some boilerplate code to quickly generate EML metadata using REML.
1.2 Speed and Rigour
As I noted in a previous post, there are [two types of data documentation workflow](http://ivanhanigan.github.io/2013/10/two-main-types-of-data-documentation-workflow/).
- GUI
- Programatic
I also think there are two types of users with different motivations and constraints:
- 1) Data Analysts
- 2) Data Librarians
1.3 Analysts can often trade-off completeness of documentation for speed
In my view the Analysts group of users need a tool that will very rapidly document their data and workflow steps and can live with a bit less rigour in the quality of documentation. Obviously this is not ideal but seems an inevitable trade-off needed to enable analysts to keep up the momentum of the data processing and modelling without getting distracted by tedious (and potentially unnecessary) data documentation tasks.
1.4 Librarians produce gold plated documentation and can take longer to produce this
On the other hand the role of the Librarian group is to produce documentation to the best level possible (given time and resource constraints) the datasets and methodologies that lead to the creation of the datasets. For that group Rigour will take precedence and there will be a trade-off in terms of the amount of time needed to produce the documentation.
1.5 An example
As an example of the two different groups, an analyst working with weather data in Australia may want to specify that their variable "temperature" is the average of the daily maxima and minima, but might not need to specify that the observations were taken inside a Stevenson Screen, or even if they are in Celsius, Farenhiet or Kelvin. They will be very keen to start the analysis to identify any associations between weather variables and the response variable they are investigating. The data librarian on the other hand will be more likely to need to include this information so that the users of the temperature data do not mis-interpret it.
1.6 Embracing Inaccuracy and Incompleteness
- I've been talking about this for a while got referred to this document by Ben Davies at the ANUSF
[http://thedailywtf.com/Articles/Documentation-Done-Right.aspx](http://thedailywtf.com/Articles/Documentation-Done-Right.aspx)
- It has this bit:
Embracing Inaccuracy and Incompleteness
The immediate answer to what’s the right way to do documentation is
clear: produce the least amount of documentation needed to facilitate
the most understanding, and be very explicit about which documentation
is to be maintained and which is to be archived (i.e., read-only and
left to rot).
- Roughly speaking, a full EML document produced by Morpho is a bit like a whole bunch of cruft that isnt needed and gets in the way (and is more confusing)
- Whereas a minimal version Im thinking of covers almost all the generic entries providing the "minimum amount of stuff to make it work right".
1.7 Aim
- This experiment aims to speed up the creation of a minimal "skeleton" of metadata to a level that both the groups above can be comfortable with AS A FIRST STEP.
- It is assumed that additional steps will then need to be taken to complete the documentation, but the automation of the first part of the process should shave off enough time to suit the purposes of both groups
- It is an imperative that the quick-start creation of the metadata does not end up costing the documentor more time later on down the track if they need to go back to many of the elements for additional editing.
1.8 Step 1: load a simple example dataset
I've been using a [fictitious dataset from a Statistics Methodology paper by Ritschard 2006](http://ivanhanigan.github.io/2013/10/test-data-for-classification-trees/). It will do as a first cut but when it comes to actually test this out it would be good to have something that would take a bit longer (so that the frustrations of using Morpho become very apparent).
#### R Code: # func require(devtools) install_github("disentangle", "ivanhanigan") require(disentangle) # load fpath <- system.file( file.path("extdata", "civst_gend_sector_full.csv"), package = "disentangle" ) data_set <- read.csv(fpath) summary(data_set) # store it in the current project workspace write.csv(data_set, "data/civst_gend_sector_full.csv", row.names = F) ## | divorced/widowed: 33 | female:132 | primary :116 | Min. : 128.9 | ## | married :120 | male :141 | secondary: 99 | 1st Qu.: 768.3 | ## | single :120 | nil | tertiary : 58 | Median : 922.8 | ## | nil | nil | nil | Mean : 908.4 | ## | nil | nil | nil | 3rd Qu.:1079.1 | ## | nil | nil | nil | Max. :1479.4 |
1.9 Step 2 create a function to deliver the minimal metadata object
- the package REML will create a EML metadata document quite easily
- I will assume that a lot of the data elements are self explanatory and take column names and factor levels as the descriptions
1.10 remlboilerplate-code
################################################################ # name:reml_boilerplate # func if(!require(reml)) { require(devtools) install_github("reml", "ropensci") } require(reml) reml_boilerplate <- function(data_set, created_by = "Ivan Hanigan <ivanhanigan@gmail.com>", data_dir = getwd(), titl = NA, desc = "") { # essential if(is.na(titl)) stop(print("must specify title")) # we can get the col names easily col_defs <- names(data_set) # next create a list from the data unit_defs <- list() for(i in 1:ncol(data_set)) { # i = 4 if(is.numeric(data_set[,i])){ unit_defs[[i]] <- "numeric" } else { unit_defs[[i]] <- names(table(data_set[,i])) } } # unit_defs ds <- data.set(data_set, col.defs = col_defs, unit.defs = unit_defs ) #str(ds) metadata <- metadata(ds) # needs names for(i in 1:ncol(data_set)) { # i = 4 if(is.numeric(data_set[,i])){ names(metadata[[i]][[3]]) <- "number" } else { names(metadata[[i]][[3]]) <- metadata[[i]][[3]] } } # metadata oldwd <- getwd() setwd(data_dir) eml_write(data_set, metadata, title = titl, description = desc, creator = created_by ) setwd(oldwd) sprintf("your metadata has been created in the '%s' directory", data_dir) }
1.11 remlboilerplate-test-code
################################################################ # name:reml_boilerplate-test analyte <- read.csv("data/civst_gend_sector_full.csv") reml_boilerplate( data_set = analyte, created_by = "Ivan Hanigan <ivanhanigan@gmail.com>", data_dir = "data", titl = "civst_gend_sector_full", desc = "An example, fictional dataset" ) dir("data")
1.12 Results: This loads into Morpho with some errors
- Notably unable to import the data file
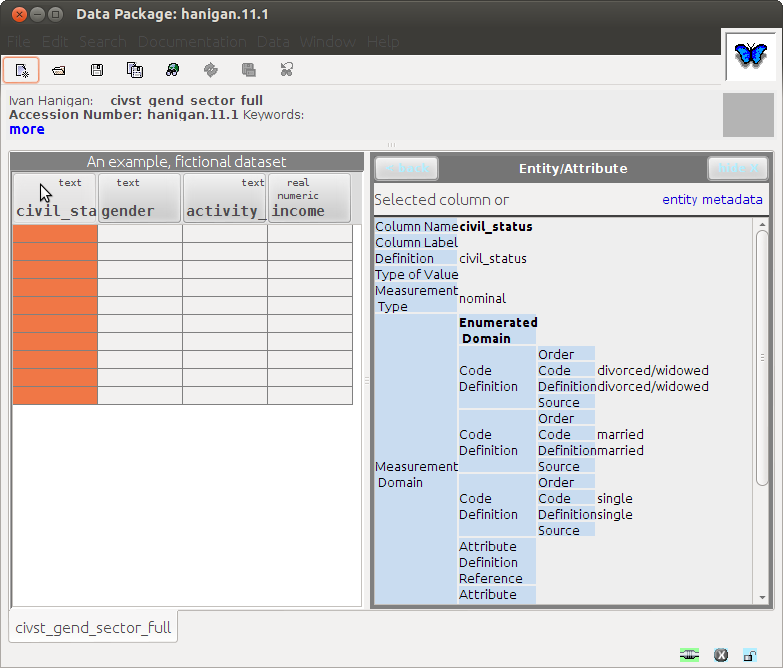
- Also "the saved document is not valid for some reason"
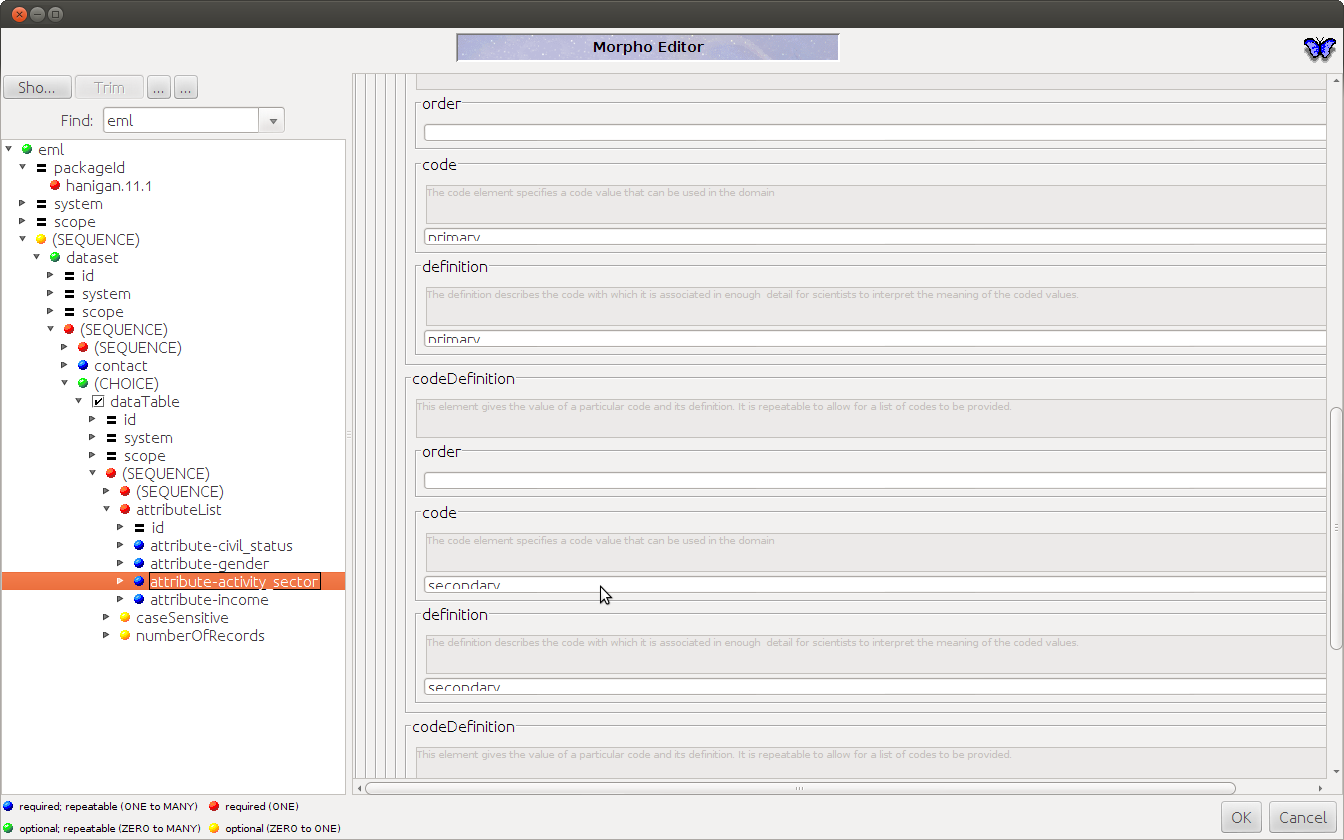
1.13 Conclusions
- This needs testing
- A real deal breaker is if the EML is not valid
- In some cases not having the data table included will be a deal breaker (ie KNB repositories designed for downloading complete data packs
- A definite failure would be that even if it is quicker to get started if it takes a long time and is difficult to fix up it might increase the risk of misunderstandings.
</html>





