Workflow flowcharts - Update
A while back I posted about my work with the Rgraphviz toolbox toward a wrapper function that will allow me to track the connections between chunks of my code as I write it. This update includes notes from a discussion I had with Keith about this.
The basic use case is described by this code:
require(disentangle)
# Decide on the first step and create the starting node of the flowchart
nodes <- newnode(name="NAME", inputs="INPUT", newgraph=T)
# FirstStep.
# Comment: do a bit of work
NAME <- myWorkFunction(INPUT)
# Decide on next step
nodes <- newnode(name="OUTPUT", inputs=c("NAME","ANOTHER THING"))
# Do the second step
OUTPUT <- mySecondFunction(NAME, ANOTHER_THING)
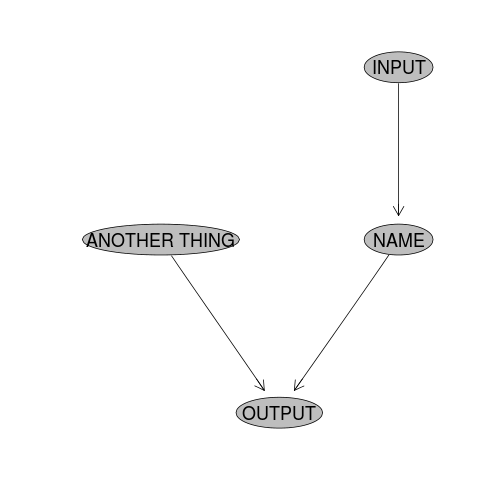
So as the scripted workflow develops, so does the flow chart. The alternative I described previously is to make a list of all the steps in a Balzerian Filelist Table.
Keith’s comments:
Keith gave me some great comments on that post. I’ll just jot them down here for now but will have to come back and re-write parts of my function and the descriptive blog post to address these at another time.
First section:
- I said that multiple datasets used in an analysis could be broken down into discrete data packages. Keith asked if I thought they “should be”.
- questioned whether the data warehouse at my work really is “Big Data”
- I need to cite the graphviz and Rgraphviz software better
- the function currently requires users to speciify newgraph = T to start, and fails to do anything if “nodes” object doesn’t exist. This is uneccessary. should make it create “nodes” if doesn’t exist. Then the newgraph argument is only needed to delete an old graph and start again.
- is it required that any of these actually be new? if not then “newnode” seems a poor name for the function
From section “Code:adding nodes”:
- the line where output is the name is confusing. Keith felt that output was not the name of the newnode (but it really is).
- the name of the nodes object is hardcoded to be “nodes”. therefore don’t need to give it as an argument
- proposed better syntac: newnode(name= “ANOTHER THING”, output = “OUTPUT”) which I think will also work?
- wouldn’t it be great if links could be labelled? maybe better to focus on links rather than nodes? this seems more logical because links are transitions and where the action is, not at the nodes.
- it might be helpful to review common actions to see what graph structures each implies.
- RE lab-book from Chemistry: most people would not be familiar with how lab-books are written.
- can newnode build circuits/cycles? can it add links between existing nodes?
Section after “shows this result”
- tablexyz -> FileA -> Input1 should be one step because fileA is not used again. ditto fileB.
- analysisResults is a different kind of thing. this is confusing.
- why do I have to type “nodes <-“ every time? if it has to have this name It could be hidden
Under example two, “Step three”
-
huh? does input1 have “id”?
-
“the result” graph is a bit dense and hard to follow. Perhaps simpler labels? ie instead of FileA just “A”?