- I recently reviewed a tool for collecting metadata about Long Term Ecological Resarch (LTER) data
- it is just an Excel spreadsheet called cwt_data_subm_template_2013.xls
- You can download a copy here www.coweeta.uga.edu/resources/forms/cwt_data_subm_template_2013.xls
- LTER is The U.S. Long-Term Ecological Research (LTER) network
- I made the following notes, this is not intended to be a nasty critique
- The following is a few Frank and Fearless comments I’ll be using to compare the pros and cons of a variety of data documentation approaches
Critique
- opened first on windows, saw comments on cells with instructions
- opened next on linux with libreOffice and comments are gone
- opened at the last tab (split in two for no reason?)
- noticed recommended name “GCE site” = Site, otherwise “permanent plot” = Plot?
- GCE = Georgia Coastal Ecosystems LTER program
- flip to first tab, point 4 suggests there is some export functionality I cannot see (a VBA script?)
- cell 11 a NOTE: When submitting updated metadata or re-using templates please highlight fields with modified contents in yellow
- and use glitter pen??? (See this great post called excel-is-not-your-lab-notebook!
- personnell tab OK
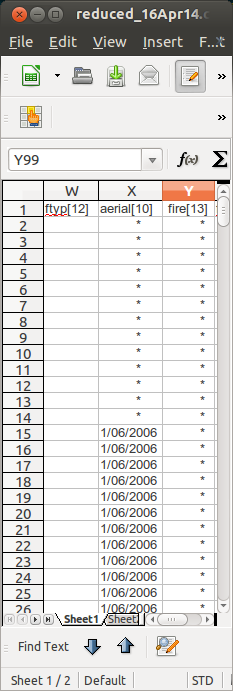
- instrumentation, variable measured is free text. ok but for eg “max temp”, “temperature maxima”, “maximum temperature (c)” “maximum temperature in 24 hours after 9am local time in degrees” etc
- too wide, last column was off my wide screen! noticed wasted real estate in column A
Moving on to the tabular data sheet
- I don’t like this “– Paste or enter your data values into the ‘Values’ section (white cells), starting with the indicated cell”
- this is an invitation for clerical error! Too many “copy-and-paste” actions will inevtably introduce errors
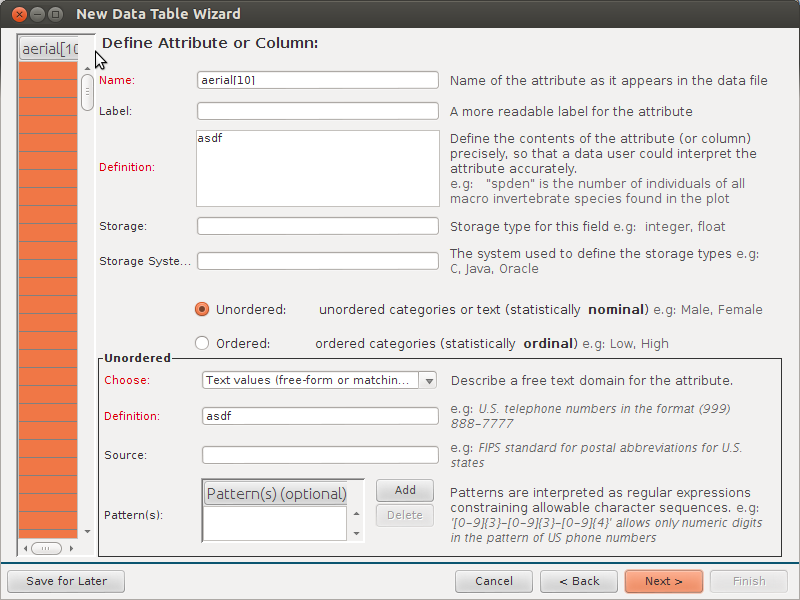
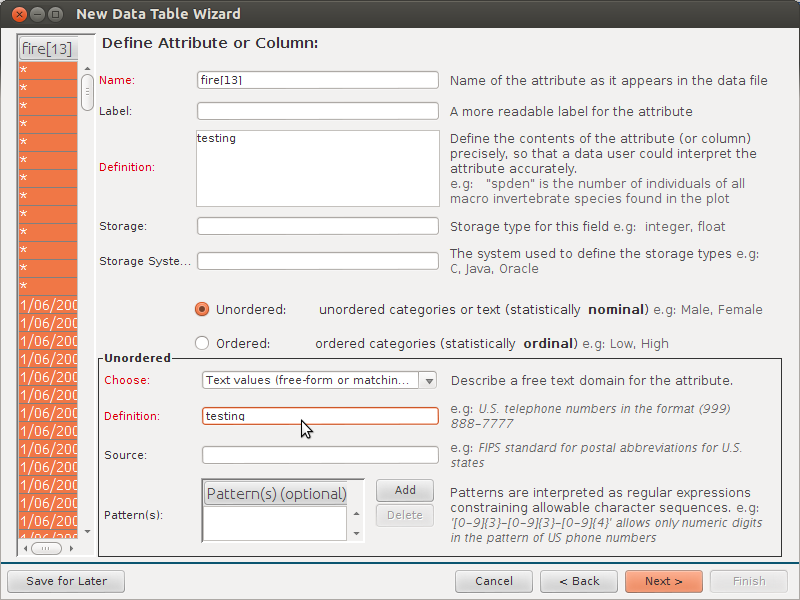
I do like the extra metadata Column Name:
- Description:
- Units:
- Data type:
- Variable type:
- Number type:
- Precision:
- Code values:
- Calculations:
- QC: Minimum Valid:
- QC: Minimum Expected:
- QC: Maximum Expected:
- QC: Maximum Valid:
- QC: Custom:
Other issues:
– Fill in missing values in the table with NaN (not a number), including text fields, and do not skip columns
- but what about missing values imbued with other meanings (NA = not observed, censored etc)?
- ask users to format digit rounding in Excel?? oh no
- old excel users may still be restricted to 65,536 rows by 256 columns.
- non tabular sheet is ok