r-eml
- this is the R package for authoring EML
pgis
- This is a spatial database
System Description
- Ubuntu 12.04 LTS
- Macpro Tower
EML is a useful beast
setting-up-the-software-stack
- First I needed devtools and this required RCurl linux package dependencies
Code
sudo apt-get install libcurl libcurl-devel libcurl4-gnutls-dev
- Now trying to install EML
Code
require(devtools)
install_github("EML", "ropensci")
- then XML Depends
Code
sudo apt-get install libcurl4-openssl-dev libxml2-dev
sudo apt-get install r-cran-xml
- rJava can be difficult, I was really dreading this but eventually stackoverflow to the rescue!
Code
sudo apt-get install openjdk-7-*
sudo apt-get install r-cran-rjava
update-java-alternatives -l
sudo update-java-alternatives -s java-1.7.0-openjdk-amd64
sudo R CMD javareconf
R
install.packages("rJava")
library(rJava)
- now complaining about ‘RHTMLForms’ and ‘RWordXML’
Code:
install_github("RHTMLForms", "omegahat")
install.packages("RWordXML", repos="http://www.omegahat.org/R", type="source")
- on we go
Code
install.packages(c("knitr", "rfigshare", "testthat", "RCurl", "dataone", "rrdf"))
RGDAL
- Spatial toolbox
Code
sudo apt-get update
sudo apt-get upgrade
sudo apt-get install libgdal1-dev libproj-dev
gdal-config --version
sudo R
install.packages("rgdal")
PostgreSQL 9.3
- following comes from http://technobytz.com/install-postgresql-9-3-ubuntu.html
Code:
sudo apt-get update
sudo apt-get -y install python-software-properties
wget --quiet -O - https://www.postgresql.org/media/keys/ACCC4CF8.asc | sudo apt-key add -
sudo sh -c 'echo "deb http://apt.postgresql.org/pub/repos/apt/ precise-pgdg main" >> /etc/apt/sources.list.d/postgresql.list'
sudo apt-get update
sudo apt-get install postgresql-9.3 pgadmin3
sudo -u postgres psql
create role user_name login password 'passwd';
show hba_file ;
#/etc/postgresql/9.3/main/pg_hba.conf
\q
sudo nano /etc/postgresql/9.3/main/pg_hba.conf
# add users to listen for at the firewall
sudo nano /etc/postgresql/9.3/main/postgresql.conf
# listenaddresses = *
sudo services postgresql restart
sudo ufw allow from my.ip.address.0/24 to any port 5432
PostGIS 2.0
- following comes from http://technobytz.com/install-postgis-postgresql-9-3-ubuntu.html
Code:
sudo apt-get update
sudo apt-get install postgresql-9.3-postgis-2.1 -f
sudo su
su - postgres
createdb postgis_ltern
psql -d postgis_ltern -U postgres
CREATE EXTENSION postgis;
CREATE EXTENSION postgis_topology;
CREATE ROLE public_group;
CREATE ROLE ivan_hanigan LOGIN PASSWORD 'password';
GRANT public_group TO ivan_hanigan;
grant usage on schema public to public_group;
GRANT select ON ALL TABLES IN SCHEMA public TO public_group;
grant execute on all functions in schema public to public_group;
grant select on all sequences in schema public to public_group;
grant select on table geometry_columns to public_group;
grant select on table spatial_ref_sys to public_group;
grant select on table geography_columns to public_group;
grant select on table raster_columns to public_group;
grant select on table raster_overviews to public_group;
For transforming AGD 66 to GDA94
- A special transformations grid file is required to be added to the PROJ.4 files for reprojecting the Australian projections AGD66 to GDA94.
- Thanks to Joe Guillaume and Francis Markham for providing this solution.
Code: transformations grid for Australian projections
cd /usr/share/proj
wget http://www.icsm.gov.au/gda/gdatm/national66.zip
unzip national66.zip
mv "A66 National (13.09.01).gsb" aust_national_agd66_13.09.01.gsb
su - postgres
psql -d mydb
UPDATE spatial_ref_sys SET
proj4text='+proj=longlat +ellps=aust_SA +nadgrids=aust_national_agd66_13.09.01.gsb +wktext'
where srid=4202;
\q
exit
make a spatial dataset
Code
require(devtools)
install_github("swishdbtools", "swish-climate-impact-assessment")
require(swishdbtools)
require(ggmap)
ch <- connect2postgres2("postgis_ltern")
location_names <- c("linnaeus way acton canberra act", "biology place acton canberra act")
locations <- geocode(location_names)
locations <- cbind(location_names, locations)
dbWriteTable(ch, "anu_gisforum_locations", locations, row.names = F)
sql <- points2geom("public", "anu_gisforum_locations", col_lat = "lat", col_long = "lon")
cat(sql)
dbSendQuery(ch,
sql
)
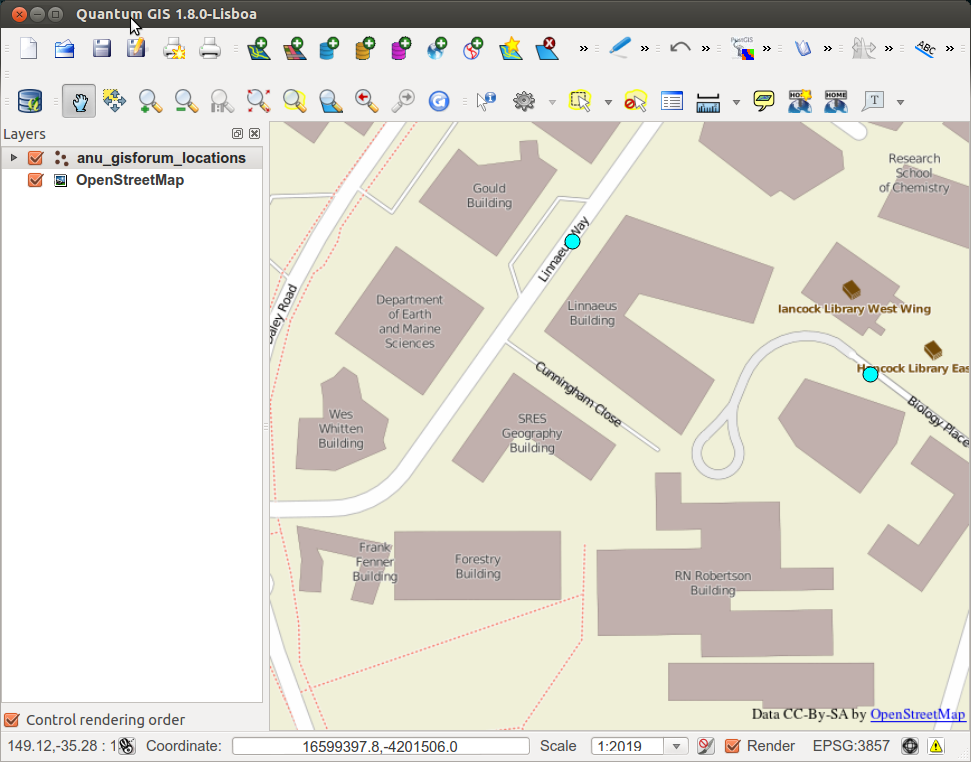
This can now be shown in Quantum GIS
TODO:
- add EML for this using R
- finesse with Morpho (requires further Java shennanigans)
- publish something to KNB using R dataone package