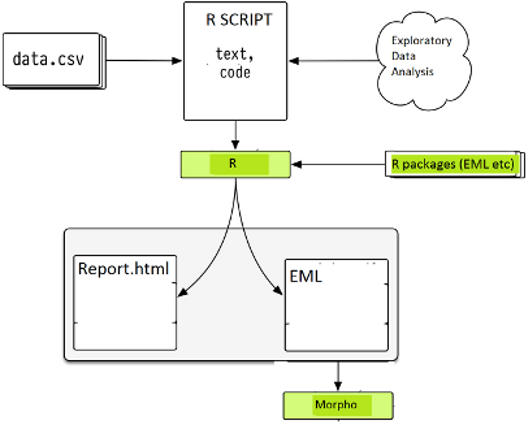
library(disentangle)
library(sqldf)
library(lubridate)
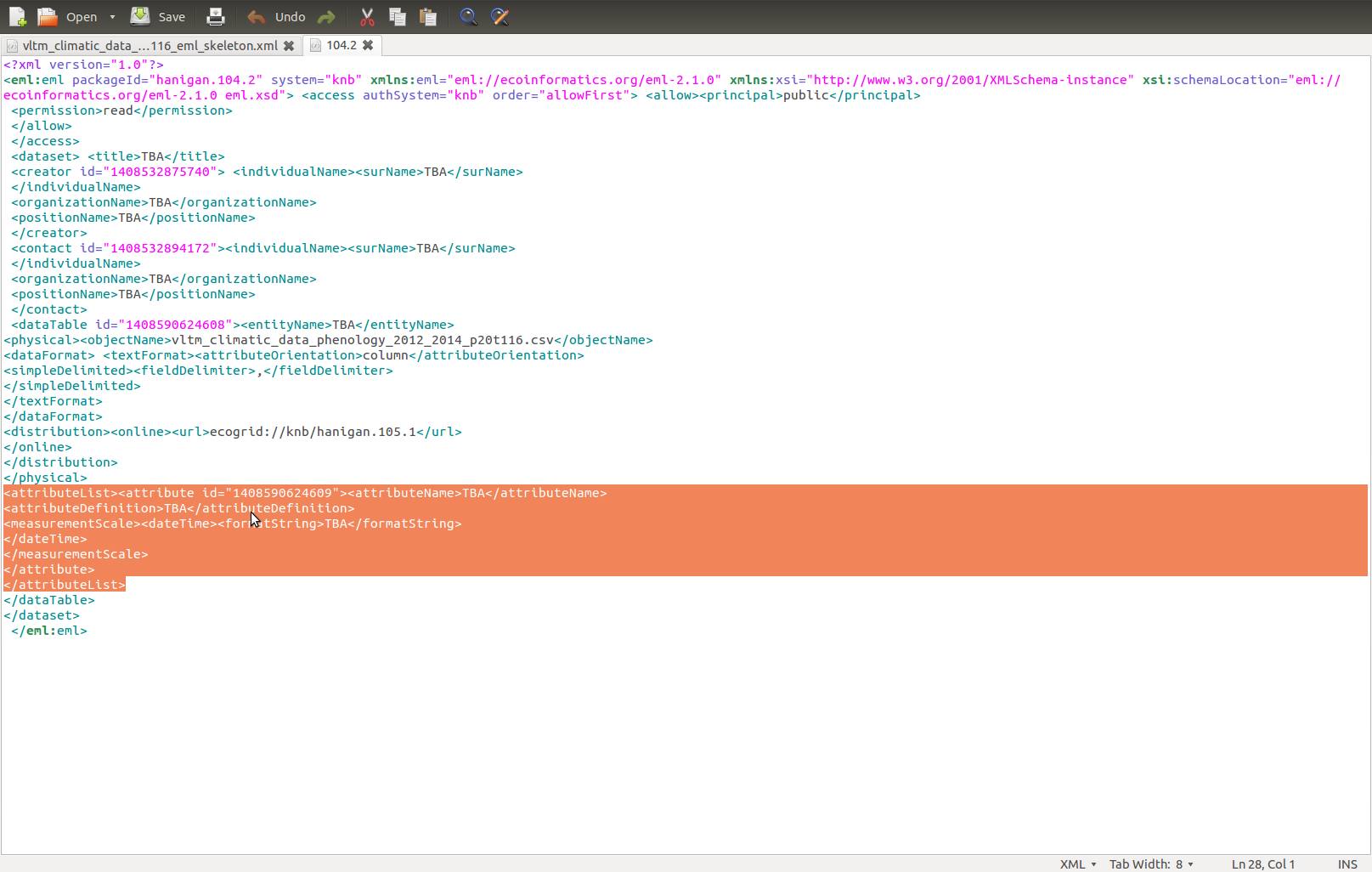
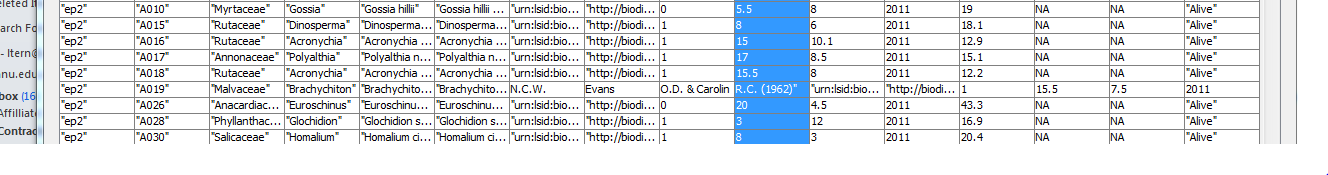
datin <- read.csv(
textConnection("
container_task_title , task_id , allocated , fte , blocker , start_date , effort , status , notes
01 Start , Start , ivan , 1 , NA , 2015-03-15 , 1d , DONE , NA
02 Update Lit Review , Repeat MEDLINE search , ivan , 1 , Start , 2015-03-16 , 5d , DONE , NA
02 Update Lit Review , Retrieve articles , ivan , 1 , Repeat MEDLINE search , NA , 5d , DONE , NA
02 Update Lit Review , Read articles , ivan , 1 , , 2015-03-26 , 11d , DONE ,
02 Update Lit Review , Summarize articles , ivan , 1 , , 2015-04-06 , 9d , TODO ,
03 Write Draft , Write introduction , ivan , 1 , , 2015-04-09 , 6d , TODO ,
03 Write Draft , Write methods , ivan , 1 , Start , , 15d , TODO ,
03 Write Draft , Write results , ivan , 1 , , 2015-03-30 , 10d , TODO ,
03 Write Draft , Write discussion , ivan , 1 , , 2015-04-15 , 10d , TODO ,
04 Internal Review , Send to co-author for review , ivan , 1 , Write discussion , , 2d , TODO ,
04 Internal Review , Revise draft 1 , ivan , 1 , , 2015-04-19 , 10d , TODO ,
05 Peer Review , Submit article 1 , ivan , 1 , Revise draft 1 , , 5d , TODO ,
06 Revise and Resubmit, Revise draft 2 , ivan , 1 , , 2015-04-30 , 10d , TODO ,
06 Revise and Resubmit, Submit article 2 , ivan , 1 , Revise draft 2 , , 5d , TODO ,
07 End , Accepted , ivan , 1 , , 2015-05-15 , 1d , TODO ,
"),
stringsAsFactor = F, strip.white = T)
# or
# datin <- get_gantt_data("gantt_todo", test_data = T) # need to
# adjust min_context_xrange to 2015-01-01 or something
datin$start_date <- as.Date(datin$start_date)
str(datin)
datin
dat_out <- gantt_data_prep(dat_in = datin)
str(dat_out)
dat_out
svg("tests/gantt_tufte_test.svg",height=10,width=8)
gantt_tufte(dat_out, focal_date = "2015-04-13", time_box = 3*7,
min_context_xrange = "2015-03-16",
cex_context_ylab = 0.65, cex_context_xlab = .7,
cex_detail_ylab = 0.9, cex_detail_xlab = .4,
show_today = F)
dev.off()