This will be a series of three posts that describe some key evidence based best practice methods that have helped me plan and organise files and folders for data analysis. I have found these via books and on websites.
- Scott Long’s Workflow for Data Analysis with Stata
- Josh Reich’s Least Commonly Fouled up Data analysis (LCFD) framework
- John Myles White’s ProjectTemplate
Toward evidence based best-practice data management systems
It is important for open science to have effective management of digital assets across the different phases of the research pipeline. The traditional research pipeline moves from steps of hypothesis and design, measured data, analytic data, computational results (for figures, tables and numerical results), and reports (text and formatted manuscript). Reproducible research pipelines extend traditional research by encoding the steps in code from a computer ‘scripting’ language, and distributing the data and code with publications.
In this research pipeline context there are a large number of potential ways to manage digital assets (documents, data and code). There are also many different motivating drivers that will affect the way that a scientist or group of scientists choose to manage their data and code.
To deal with in house data management issues before starting and during analysis/reporting is critical for reproducible research.
I argue that more effective research pipelines can be achieved if scientists adopt the ‘convention over configuration’ paradigm and adopt best-practice systems based on evidence.
Long, S. (2015). Workflow for Reproducible Results.
For ages I was aware of the book from the Stata statistical program publishers http://www.indiana.edu/~jslsoc/web_workflow/wf_home.htm:
Citation:
Long, J. S. (2008). The Workflow of Data Analysis:
Principles and Practice. Stata publishing.
Recently I stumbled across more recent workshop slides and tutorial material which I will discuss briefly.
Citation:
Long, S. (2015). Workflow for Reproducible Results.
IV : Managing digital assets Workflow for Tools for your WF.
Retrieved from http://txrdc.tamu.edu/documents/WFtxcrdc2014_4-digital.pdf
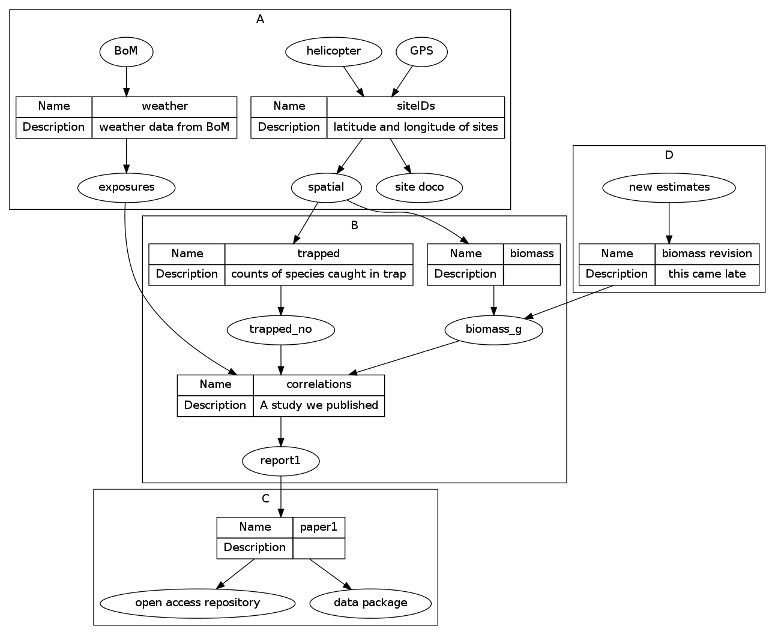
Long suggests a lot of practical things to do, but I will just focus here on the recommended file and folder structure:
Recommended project directory structure:
\ProjectAcronym
\- History starting YYYY-MM-DD
\- Hold then delete
\Admin
\Documentation
\Posted
\Paper 1
\Correspondence
\Text
\Analysis
\PrePosted
\Resources
\Write
\Work
- In another workshop report Long provides a useful tool to automatically create this structure on windows
- Long, S. (2012). Principles of Workflow in Data Analysis. Retrieved from http://www.indiana.edu/~wim/docs/2012-long-slides.pdf
- a bash version would be useful for linux and mac users, but also the R language can do this on all platforms with the
dir.createcommand
Code: wfsetupsingle.bat
# wfsetupsingle.bat
REM workflow talk 2 \ wfsetupsingle.bat jsl 2009-07-12
REM directory structure for single person.
FOR /F "tokens=2,3,4 delims=/- " %%a in ("%DATE%") do set CDATE=%%c-%%a-%%b
md "- History starting \%cdate%"
md "- Hold then delete "
md "- Pre posted "
md "- To clean"
md "Documentation"
md "Posted"
md "Resources"
md "Text\- Versions\"
md "Work\- To do"
Critical reflections
- This recommendation is very sensible, especially the suggestion of moving things through the pipeline as they evolve from things being worked on (Write/Work) to later phases when they have been polished to a point that they can be put down while preparations for distrubuting them are made (Preposted) and then once they are sent off into downstream publication phases (Posted) they are locked for ever in a archival state.
- I am not particularly keen on the names that have been chosen (Resources, Write and Work are quite ambiguous terms).