My work yesterday on implementing a lit review section of my data inventory database went pretty well, and I tested this while collating information on the papers I compile into my thesis.
However, as I worked through the information for each paper I realised that attaching this stuff at the EML/dataset level is not always going to work. In particular I have projects in which several papers are part of the one dataset. To deal with this I have invented a non-EML relationship module for publications. So in my new schema the following is possible
eml/project/
/dataset1/
/publication1
/publication2
/dataset2/
/etc
In this scenario a single dataset (ie bushfire smoke, temperature and mortality) can be used to write one paper focused on smoke, controlling for temperature. Another paper on heatwaves, controlling for smoke. Indeed we might use time-series Poisson models for one and case-crossover design for the other (this is similar to what I did with Johnston and Morgan).
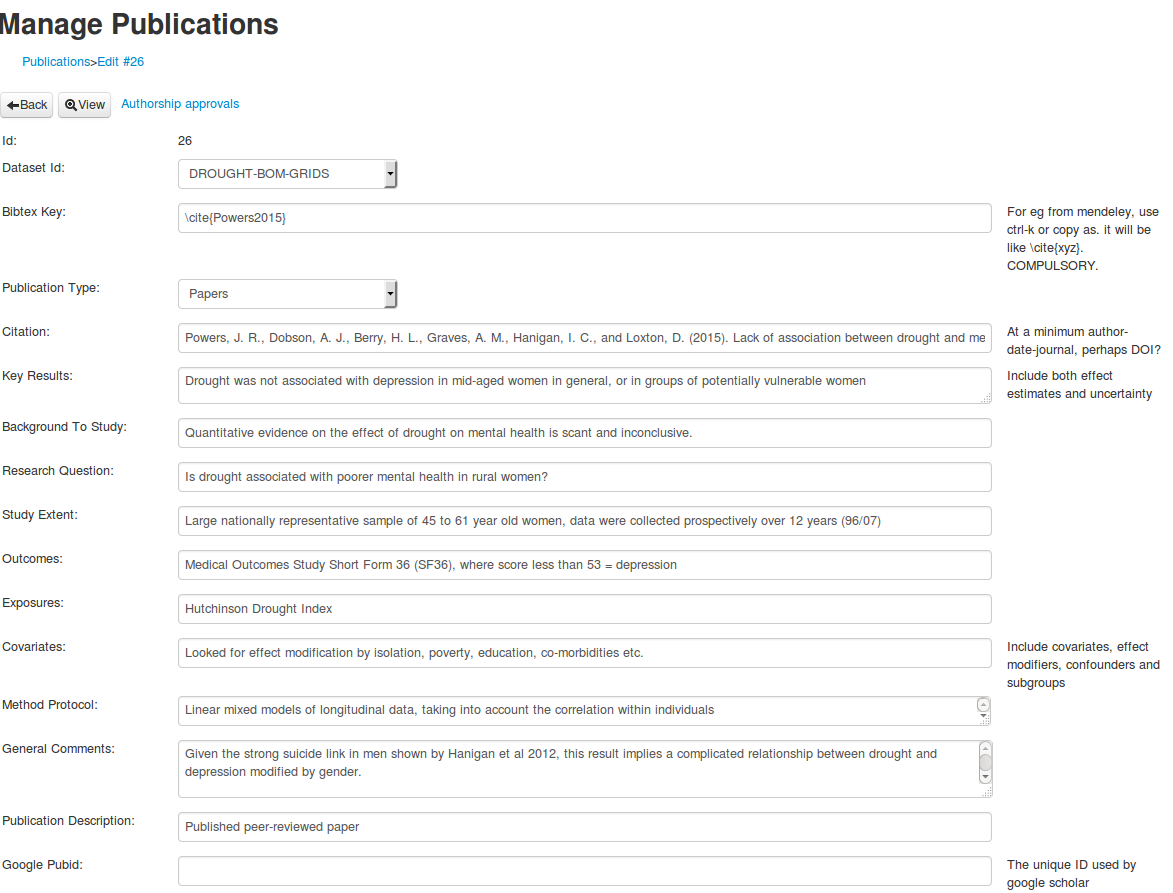
So the simple thing to do is input these ‘evidence tables’ fields at the publication level, rather than the dataset as I did yesterday.
This also partially solves my problem about aggregating these non-EML tags inside the text of the abstract, and worrying about parsing that to extract the elements of information.
The fields
| data_inventory_field | description |
|---|---|
| Citation | At a minimum author-date-journal, perhaps DOI? |
| Key results | Include both effect estimates and uncertainty |
| Background to the study | |
| Research question | |
| Study extent | |
| Outcomes | |
| Exposures | |
| Covariates | Include covariates, effect modifiers, confounders and subgroups |
| Method protocol | |
| General Comments |
An example