- In my previous post I reviewed a paper by Noble 2009 that proposed recommendations for best practice ways to set up a data analysis pipeline http://ivanhanigan.github.io/2015/10/a-quick-review-of-a-quick-guide-to-organizing-computational-biology-projects/
- I was following up on a series of posts I made about other best practice recommendations http://ivanhanigan.github.io/2015/09/reproducible-research-and-managing-digital-assets-part-3/
- In the paper by Noble it is suggested that a one should use a ‘driver script’ to automate creation of a directory structure, this is the exact way that
ProjectTemplateandmakeProjectwork as I described them in the series of posts. - I think Noble’s framework offers something new to the recomendations I had canvassed, that is the idea of chronological order of the contents of the results directory. I think this is an eminently sensible idea and thought that the R function
Sys.Date()would be a great way to start off a project in this way. - so I have put together the following R function, as an alternative to the
makeProjectcore function, that I thought I’d name so that there may be a family of makeProject functions, so that analysts have a range of to choose from. The other candidate would bemakeProjectLong, which I will also put up before long.
makeProjectNoble <- function(rootdir = getwd()){
if(!exists(rootdir)) dir.create(rootdir)
dir.create(file.path(rootdir,'doc'))
dir.create(file.path(rootdir,'doc','paper'))
sink(file.path(rootdir,'doc','workplan.Rmd'))
cat(sprintf("---\ntitle: Untitled\nauthor: your name\ndate: %s\noutput: html_document\n---\n\n",
Sys.Date()))
sink()
dir.create(file.path(rootdir,'data'))
dir.create(file.path(rootdir,'data', Sys.Date()))
dir.create(file.path(rootdir,'src'))
dir.create(file.path(rootdir,'results'))
dir.create(file.path(rootdir,'results', Sys.Date()))
file.create(file.path(rootdir,'README.md'))
}
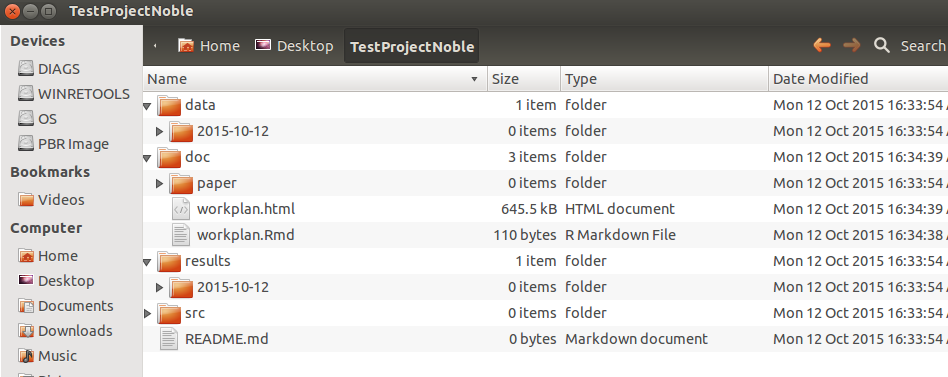
- Running this function will deploy the folders and files (I excluded the
binfolder for compiled binaries, as I believe that many data analysts may not need that, and those who do are geeky enough to write their own driver scripts.
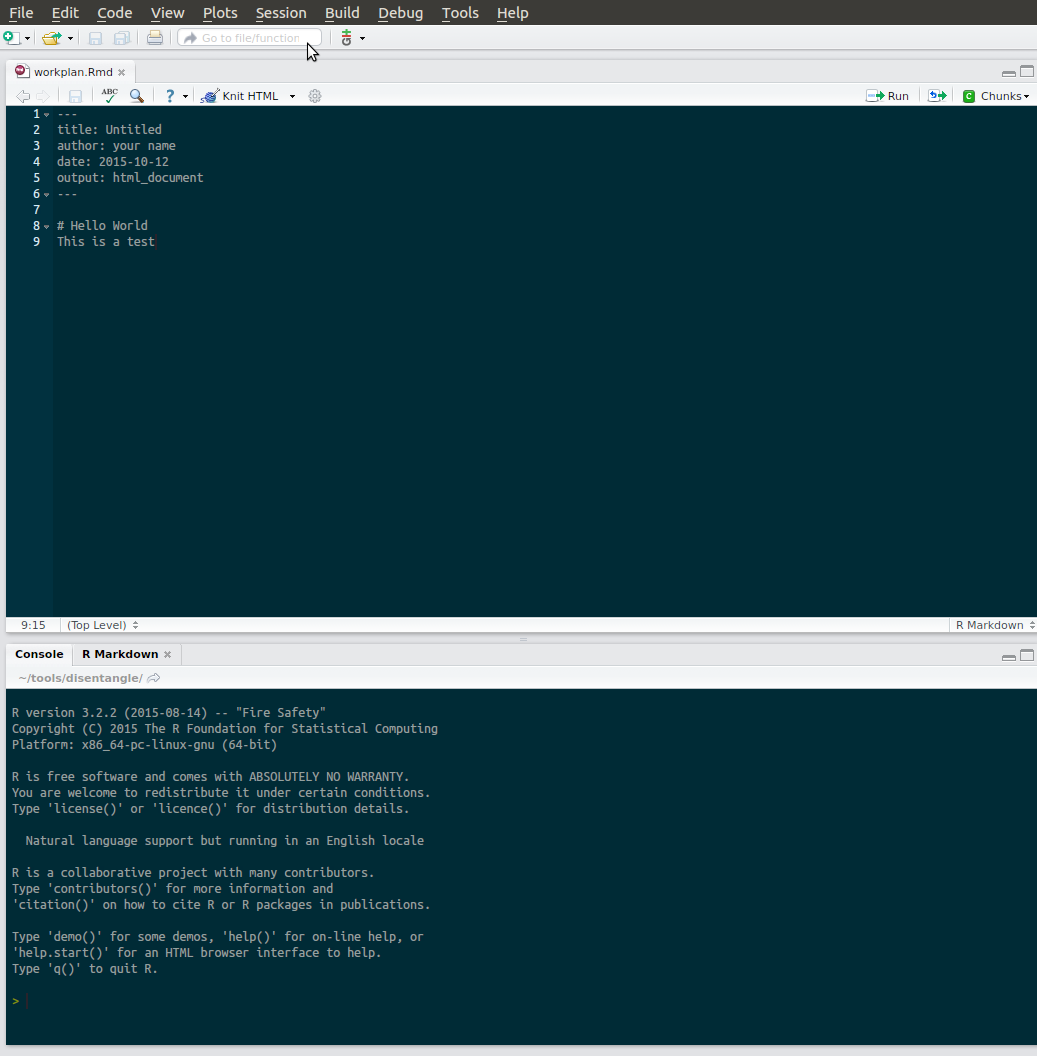
- The Rmarkdown script is waiting for the analysis plan to be pumped out, and work can begin
References
- Noble, W. S. (2009). A quick guide to organizing computational biology projects. PLoS Computational Biology, 5(7), 1–5. http://dx.doi.org/10.1371/journal.pcbi.1000424





