I use the R package DiagrammeR for creating graphs (the formal kind, connecting nodes with edges)
- This package is great and I like how it interacts with the Graphviz program
- One thing that I like to do in planning and organising data analysis projects is to make graphs and lists of the methods steps, inputs and Outputs
- A simple way to organise these things is in a dataframe (table) with a column for each step (node) and two others for inputs and outputs (edges)
- In my utilities R package
github.com/ivanhanigan/disentangleI have written functions that turn this table into a graphiviz DOT language script - Recently I have needed to unpack the list for a more itemized view
- Both these functions are showcased below
Code: newnode
# First create some test data, each step is a collection of edges
# with inputs or outputs simple comma seperated lists
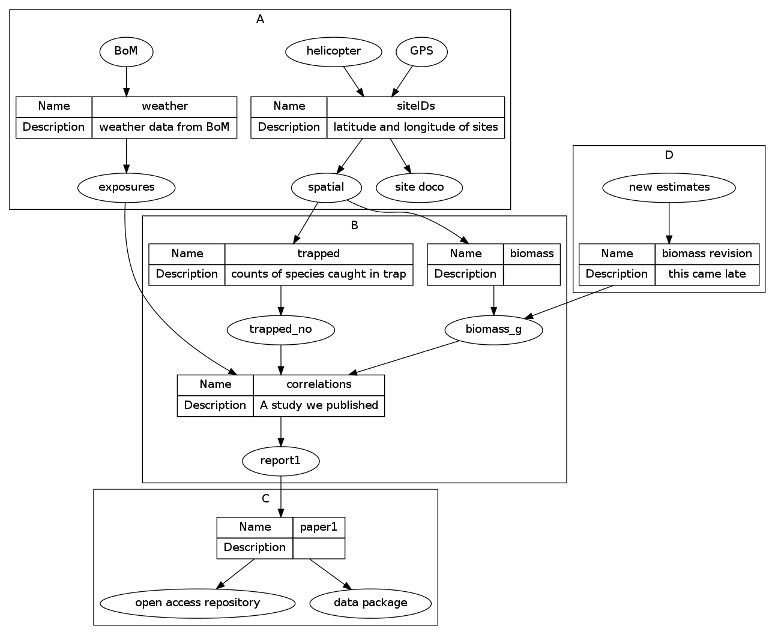
dat <- read.csv(textConnection('
cluster , step , inputs , outputs , description
A , siteIDs , "GPS, helicopter" , "spatial, site doco" , latitude and longitude of sites
A , weather , BoM , exposures , weather data from BoM
B , trapped , spatial , trapped_no , counts of species caught in trap
B , biomass , spatial , biomass_g ,
B , correlations , "exposures,trapped_no,biomass_g" , report1 , A study we published
C , paper1 , report1 , "open access repository, data package" ,
D , biomass revision, new estimates , biomass_g , this came late
'), stringsAsFactors = F, strip.white = T)
str(dat)
# dat
# Now run the function and create a graph
nodes <- newnode(
indat = dat,
names_col = "step",
in_col = "inputs",
out_col = "outputs",
desc_col = "description",
clusters_col = "cluster",
nchar_to_snip = 40
)
sink("Transformations.dot")
cat(nodes)
sink()
#DiagrammeR::grViz("Transformations.dot")
system("dot -Tpng Transformations.dot -o Transformations.png")
browseURL("Transformations.png")
That creates this diagram
Now to showcase the tool that itemizes this list of inputs and outputs
- The original table has no capacity to add detail about each node as they are held as a list of inputs and outputs
- To add detail for each we need to unpack each list and create a new table with one row per node
- I decided to make this a long table with an identifier for each node about which step (edge) the node is an input or an output
Code: newnode_csv
nodes_graphy <- newnode_csv(
indat = dat,
names_col = "step",
in_col = "inputs",
out_col = "outputs",
clusters_col = 'cluster'
)
# which creates this table
knitr::kable(nodes_graphy)
|cluster |name |in_or_out |value |
|:-------|:----------------|:---------|:----------------------|
|A |siteIDs |input |GPS |
|A |siteIDs |input |helicopter |
|A |siteIDs |output |spatial |
|A |siteIDs |output |site doco |
|A |weather |input |BoM |
|A |weather |output |exposures |
|B |trapped |input |spatial |
|B |trapped |output |trapped_no |
|B |biomass |input |spatial |
|B |biomass |output |biomass_g |
|B |correlations |input |exposures |
|B |correlations |input |trapped_no |
|B |correlations |input |biomass_g |
|B |correlations |output |report1 |
|C |paper1 |input |report1 |
|C |paper1 |output |open access repository |
|C |paper1 |output |data package |
|D |biomass revision |input |new estimates |
|D |biomass revision |output |biomass_g |
This can now be useful for making a ‘shopping list’ of the data to aquire, transform, analyse or archive.





