I will use the NMMAPSlite datasets for a simple example of what I describe as “Spatially Structured Timeseries” as opposed to “Spatio-Temporal” which I think more explicitly includes spatial structure in the model. See This Report for all the gory details.
R Codes
Spatiotemporal Regression Modelling
Table of Contents
1 Core Model
################################################################ # name:core # func setwd("~/projects/spatiotemporal-regression-models/NMMAPS-example") require(mgcv) require(splines) # load analyte <- read.csv("analyte.csv") # clean analyte$yy <- substr(analyte$date,1,4) numYears<-length(names(table(analyte$yy))) analyte$date <- as.Date(analyte$date) analyte$time <- as.numeric(analyte$date) analyte$agecat <- factor(analyte$agecat, levels = c("under65", "65to74", "75p"), ordered = TRUE ) # do fit <- gam(cvd ~ s(tmax) + s(dptp) + city + agecat + s(time, k= 7*numYears, fx=T) + offset(log(pop)), data = analyte, family = poisson ) # plot of response functions png("images/nmmaps-eg-core.png", width = 1000, height = 750, res = 150) par(mfrow=c(2,3)) plot(fit, all.terms = TRUE) dev.off()
2 Core Model Plots
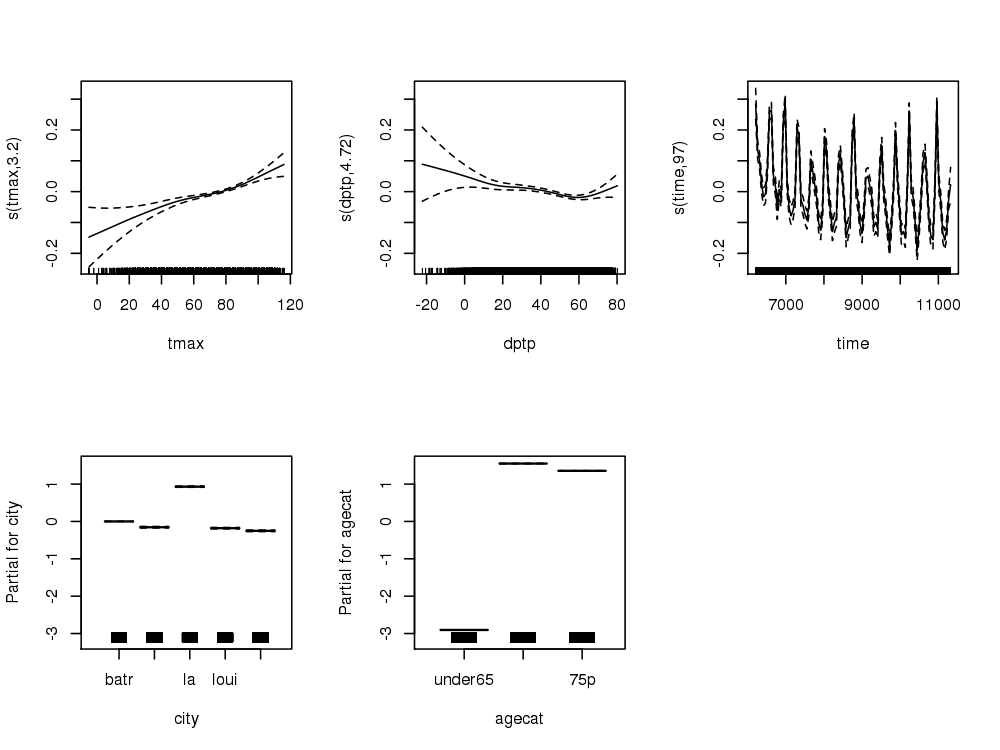
</html>





