Introduction
- at my workplace we have had a significant number of issues trying to use Morpho to publish ecological data
- I have been interested in the R EML package as an alternative and got this advice from the authors
- “I’m really happy to see you combining these tools and making best use of each of their features (authoring for Morpho, automation for REML). We should think more about how to make this combination seamless.” https://github.com/ropensci/EML/issues/93
- That dream still seems a long way off (and I suspect will see a re-write of morpho in R’s “shiny apps” language)
- but here is an attempt to circumvent some of our main issues which seem to do with using Morpho to do anything with data tables
- (morpho seems fine when just dealing with the documentation parts of EML)
Create a Minimal Morpho EML
- I am going to use Morpho to create a new data package and just step thru all the steps, add TBA to all the options
- another option was to create this using the REML example
- but then there is uncertainty if it will be accepted easily by Morpho when imported
- seriously, just TBA and only for the required fields (these are in red)
- take note of the docid number, and save locally
- now we can import this as our template. lets save it as a reference file
Code:
cp ~/.morpho/profiles/hanigan/data/hanigan/XX.1 ~/tools/morpho_template_eml.xml
Import to morpho
- this skeletal eml can now be imported in the File menu / import
- this is the ‘hotwired’ starting point of our morpho packages
- I want to do this for others too. so change morpho profile on your computer and import the xml
- gives a new package with a new morpho generated docid number
- add the package title, with enough identification in the title to allow quick reference
- SET THE ACCESS TO NOT PUBLIC BEFORE SAVING
generate the EML Skeleton using the R EML package
Code:
library(EML)
library(devtools)
install_github("disentangle", "ivanhanigan")
unit_defs <- reml_boilerplate(dat,titl = "myFile")
# you just got a quick and dirty unit_defs, these need to be made proper in morpho
# we can get the col names easily
col_defs <- names(dat)
# then create a dataset with metadata
ds <- data.set(dat,
col.defs = col_defs,
unit.defs = unit_defs
)
# now write EML metadata file
eml_config(creator="TBA <fakeaddress@gmail.com>")
wd <- getwd()
setwd(outdir)
eml_write(ds,
file = gsub(".csv", "_eml_skeleton.xml", outfile),
title = gsub(".csv", "", outfile)
)
tempfile <- dir(pattern="^data_table_")
file.rename(tempfile, outfile)
# rename the CSV file.
setwd(wd)
now attach the data
- to avoid risk of morpho getting confused it is probably safe to let it start the new dataTable tags
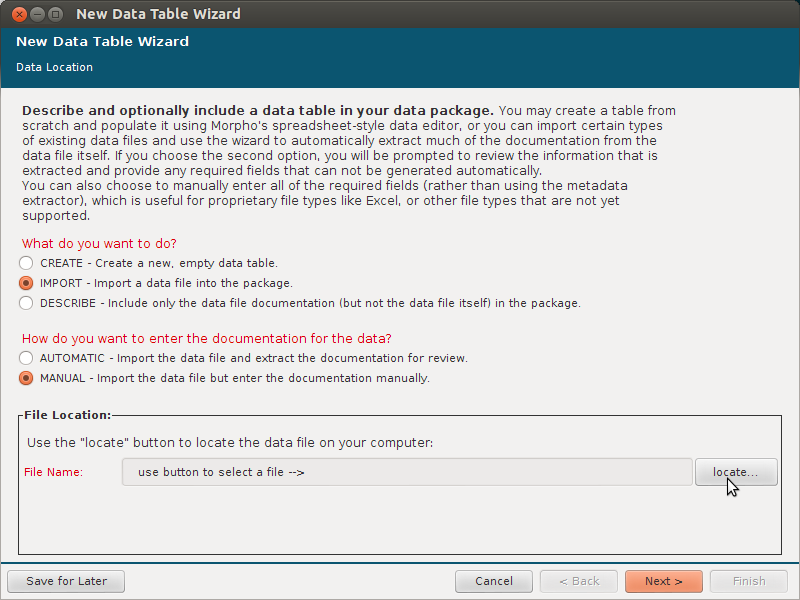
- say it is a simple CSV
- say the name for the file for title, TBA for all other required fields, for attributes add one and say it is datetime (to be quick)
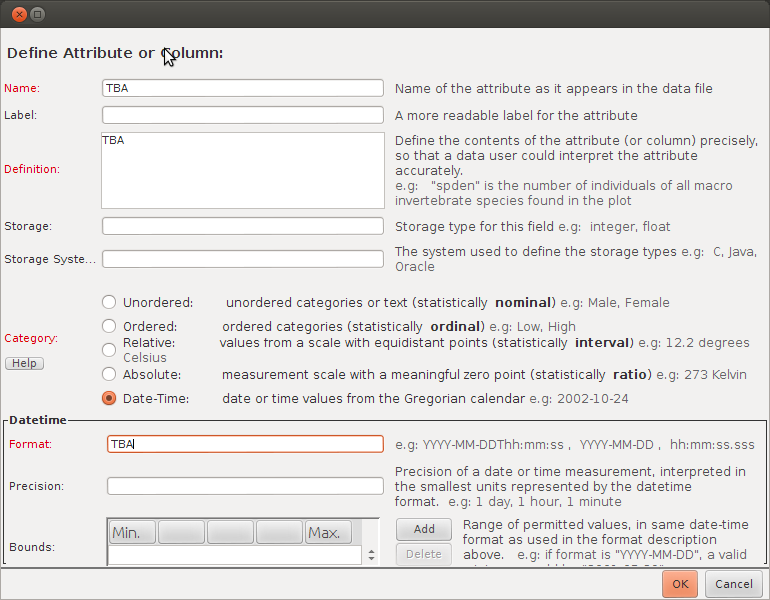
- note the docid (41.2) and download file id (ecogrid://knb/datalibrarian2.42.1)
- save locally and close
now replace the metadata info
- use an XML editor to go to the eml skeleton we created in morpho and also to the one created in R
- in the morpho generated XML find the attributeList tag within the dataTable group, mine looked like
Code:
<attributeList><attribute id="1408536821935">...
- and we know that the stuff we want to replace finishes with the closing tag </attributeList>
- I think the ID is irrelevant, except Morpho won’t allow multiple of the same id number within an EML
- find the same and replace in the REML generated bit
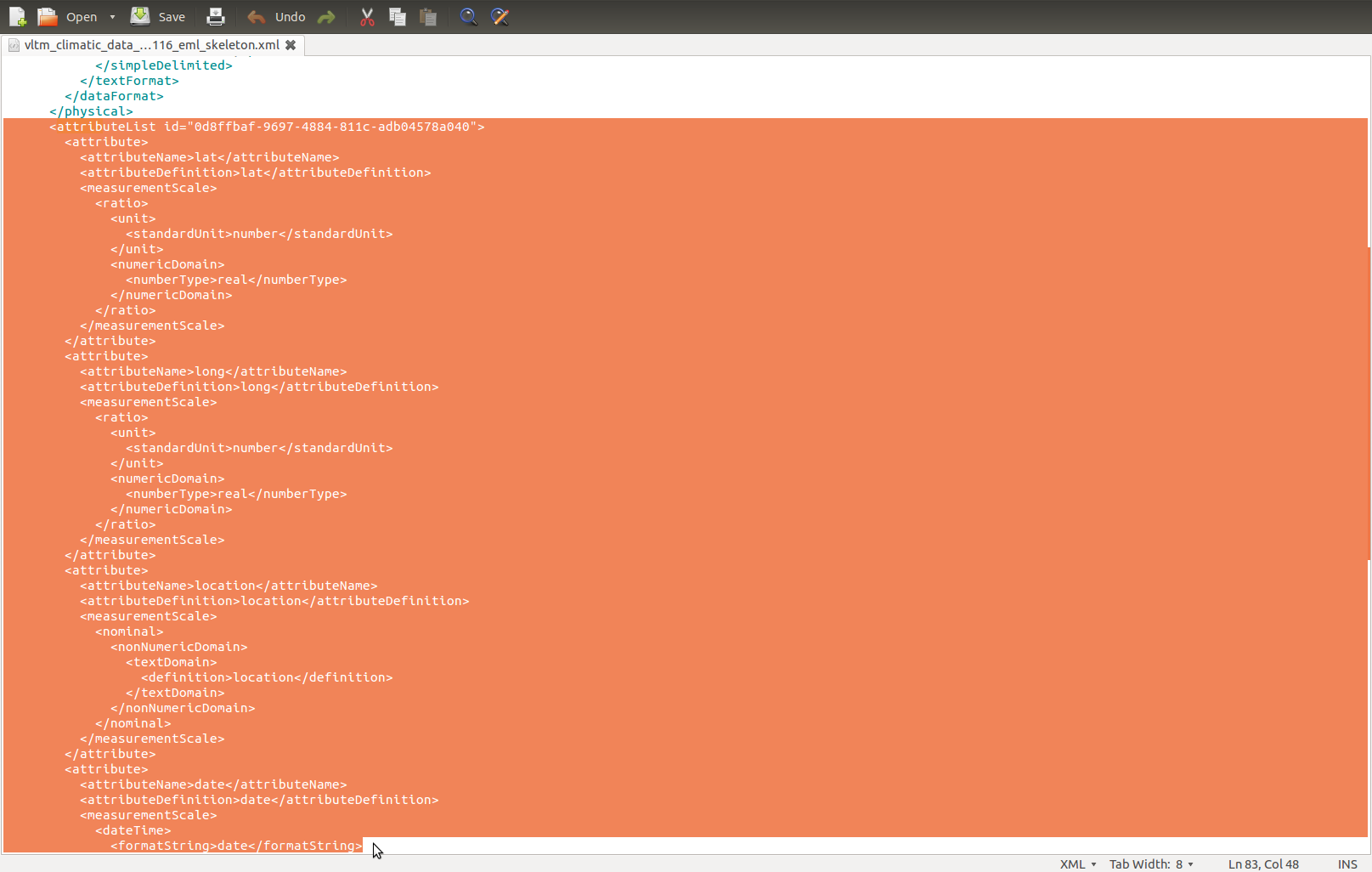
And paste:
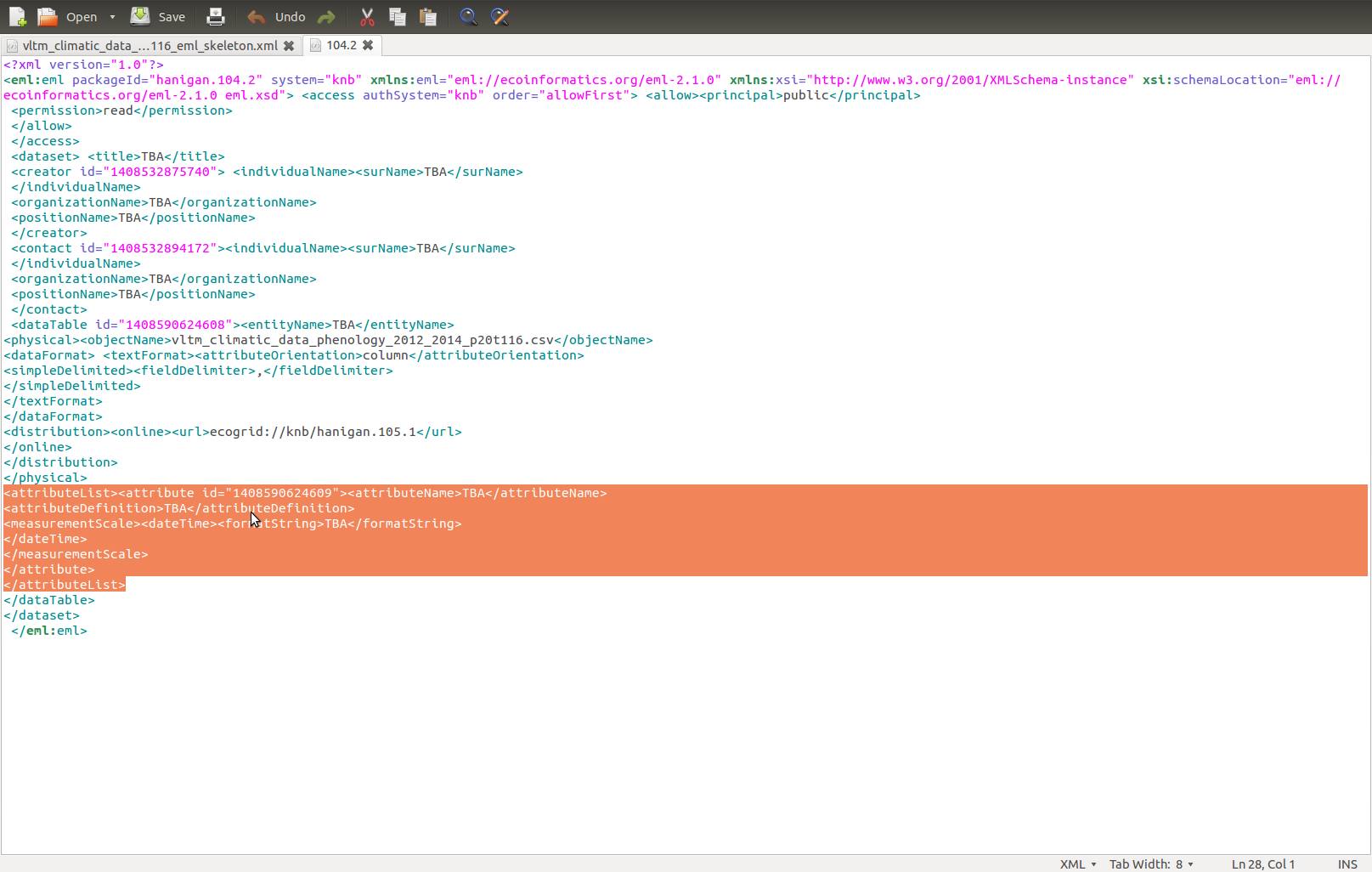
Also do the
<dataFormat></dataFormat> section
add another dataset
- assuming you already ran reml_boilerplate for this new file
- use morpho menus to add another datatabe
- save locally and close morpho
- open with XML editor and the REML EML Skeleton
- transfer dataformat and attributelist stuff
- go to documentation and make minor mod (space in abstract?), save locally
share it with colleagues
- go to documentation menu > access info and set/verify to public = no add user
- save to metacat
- now go to their (windows?) machine and log in as the same morpho profile,
- log in to the metacat and download the new package for work





