Aim
- The EML R package uses the Ecological Metadata Language (EML) approach that allows archiving of very heterogeneous data without having to standardize everything into a narrow and pre-defined syntax.
- XML files in specificed schemas involve strict criteria and are thus best generated by software.
- Morpho is an application that provides another GUI based method of generating EML but is a rather tedious tool for generating EML files. Unfortunately, without the ability to script inputs or automatically detect existing data structures, we are forced through the rather arduous process of adding all metadata annotation each time.
- The aim of this experiment is to use the EML package to create some advanced metadata quickly and then finish this off with Morpho, using “boilerplate code” wherever possible.
- this builds on my previous post http://ivanhanigan.github.io/2013/10/morpho-and-reml-streamline-the-process-of-metadata-entry
Background
- I am basically very lazy when it comes to entering metadata and when I use the Morpho package for metadata data entry I get frustrated with having to step through ever SINGLE variable and use the drop down menus etc to describe them as essentially “number” or “text”
- A big reason I like Morpho is because Metacat is a great data portal and Kepler is a promising scientific workflow tool, and all three are produced by the same group so it would be great to get them working together…
- Morpho and Metacat are open source software designed to host all kinds of ecological data. More information about it can be found here
- More info about the Metacat Data Portal System is here.
- For technical reasons, I’m running an older version of the Morpho software because I’m working with an older version of the Metacat portal software and so are also constrained to running the older Morpho version too (but will be upgrading soon).
- You might want to look at the background of the Ecological Metadata Language (EML) standard. I like this page http://carlboettiger.info/2013/06/23/notes-on-leveraging-the-ecological-markup-language.html along with the references he cites at the bottom.
Material
-
To tie this experiment back to something that is actually useful for a scientist, I will use the field-based example data on the effects on mulgara of removing spinifex, from:
McCarthy, M. a., & Masters, P. (2005). Profiting from prior information in Bayesian analyses of ecological data. Journal of Applied Ecology, 42(6), 1012–1019. doi:10.1111/j.1365-2664.2005.01101.x
-
Brief description is:
an experimental manipulation of habitat was conducted by Masters, Dickman & Crowther (2003) in which vegetation cover of a site in arid inland Australia was reduced and the response of the mammal fauna monitored.
- you can find the data in the download file from the “Code for analysing the mulgara experiment” from here
- I first checked that these data aren;t already on the KNB repository
- I searched for Marsupial, Australia, Mulgara and etc, finding no hits.
- We will assume that these data are not already published there.
Methods
- Step one install the github version of EML and a function I wrote for this a while back
- Note that I made a few changes since that post https://github.com/ivanhanigan/disentangle/blob/master/R/reml_boilerplate.r
R Code:
# func
library("devtools")
install_github("EML", "ropensci")
library("EML")
install_github("disentangle", "ivanhanigan")
library("disentangle")
# load
datatext <- 'Treat, Before, After1, After2
0, 2.833213344, 1.609437912, 2.48490665
0, 1.791759469, 2.197224577, 2.079441542
0, 3.044522438, 2.708050201, 3.135494216
0, 2.772588722, 1.791759469, 2.197224577
0, 1.098612289, 1.609437912, 2.63905733
1, 2.944438979, 0.693147181, 1.791759469
1, 2.564949357, 0.693147181, 1.791759469
1, 2.564949357, 1.609437912, 1.609437912
1, 0.693147181, 1.098612289, 1.098612289
1, 1.609437912, 0, 1.098612289'
analyte <- read.csv(textConnection(datatext))
# check
analyte
# do
## from a work dir with a subdir for data
write.csv(analyte, "data/mulgara.csv", row.names = F)
reml_boilerplate(
data_set = analyte
,
created_by = "Ivan Hanigan <ivanhanigan@gmail.com>"
,
data_dir = "data"
,
titl = "mulgara"
,
desc = "Experimental data: effect of cover reduction on mulgara Dasycercus cristicauda"
)
- Now open morpho and under file > import browse to this data directory and import.
- Got several warnings, about unable to display data, and an older version that could be updated
Results
- Result does not display in morpho
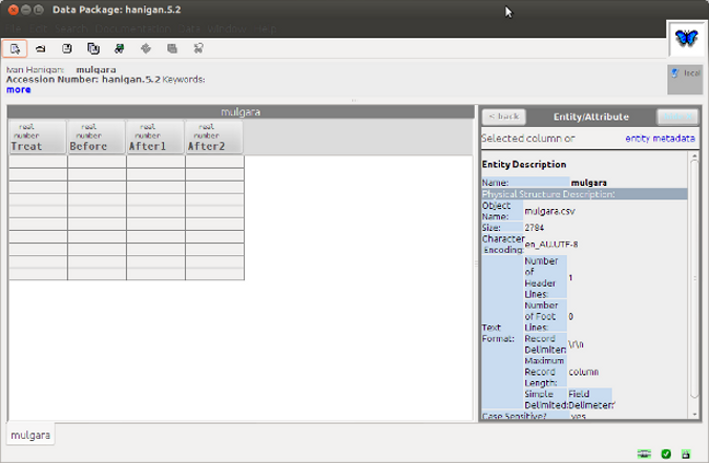
Discussion
- There is something different about the way the EML R package writes the data and what Morpho 1.8 is expecting.
Conclusions
- Further research is required.





