Spatiotemporal multilevel modelling
Table of Contents
- 1 Introduction
- 2 Compare basic regression with mixed-effects modelling
- 3 Intercepts and coefficients that vary by group
- 4 Spatially structured timeseries
- 5 Spatial lag and timeseries
1 Introduction
1.1 Overview
In this notebook we will introduce what multi-level data and models are and why they are useful in epidemiology contexts. We begin with the general linear model framework GLM and then move to mixed-effects models. It is important to realise that these models are called different things in different disciplines: (generalized) linear mixed (effects) models (GLMM), hierarchical (generalized) linear models, etc. The terminology for the model parameters is equally diverse, usually including the terms random effects and fixed effects. So we focus on the key concepts.
We will cover what is a Random Effect and how it differs from a Fixed effect. Some example syntax in R (on a secure website portal we have configured) and Stata will show how to get a handle on models that have random intercepts and additionally, random slopes.
We will spend a little time talking about how to partition variation and get estimates of the random and fixed effects. An important element will be our discussion of similarities between mixed-effects models with basic regression. There will also be brief discussion of extending the regression to non-gaussian responses (GLMERs).
We will discuss how to interpret these results in a population health context.
An online analysis server (Rstudio in a web browser) and data repository (geoserver) are also provided
2 Compare basic regression with mixed-effects modelling
2.1 Terminology
When discussing multilevel modelling in this workshop we will avoid using the names 'fixed effects' and 'random effects'. Instead we use the more verbose phrase: intercepts/coefficients that are common across groups and intercepts/coefficients that vary by group. This is because, as noted in the introduction to Gelman and Hill 2007 (recommended reading) it is very problematic that these terms are used in imprecise and confusing ways that vary across disciplines. This concern also appears in the work of the rstanarm package developers and in the vignette 'Estimating Generalized Linear Models with Group-Specific Terms with rstanarm' who state:
'Models with this structure are referred to by many names: multilevel models, (generalized) linear mixed (effects) models (GLMM), hierarchical (generalized) linear models, etc. The terminology for the model parameters is equally diverse… [and commonly used terms] are not only misleading but also defined differently across the various fields in which these models are applied.'
2.2 Introduce the linear model and lmer
3 Intercepts and coefficients that vary by group
3.1 Ross River virus and urban vs rural mosquito habitat
See domodelcheckingglmer
#########################
4 Spatially structured timeseries
I will use the NMMAPSlite datasets for a simple example of what I describe as "Spatially Structured Timeseries" as opposed to "Spatio-Temporal" which I think more explicitly includes spatial structure in the model.
4.1 Outcome Data
4.1.1 Original Data
" http://cran.r-project.org/web/packages/NMMAPSlite/index.html Package ‘NMMAPSlite’ was removed from the CRAN repository. Formerly available versions can be obtained from the archive. Archived on 2013-05-11 at the request of the maintainer. The Archived versions do not seem to work either. That is such a shame, lucky I saved some of the data using the following code: "
#### Code: get nmmaps data # func if(!require(NMMAPSlite)) install.packages('NMMAPSlite');require(NMMAPSlite) ###################################################### # load setwd('data') initDB('data/NMMAPS') # this requires that we connect to the web, # so lets get local copies setwd('..') cities <- getMetaData('cities') head(cities) citieslist <- cities$cityname # write out a few cities for access later for(city_i in citieslist[sample(1:nrow(cities), 9)]) { city <- subset(cities, cityname == city_i)$city data <- readCity(city) write.table(data, file.path('data', paste(city_i, '.csv',sep='')), row.names = F, sep = ',') } # these are all tiny, go some big ones for(city_i in c('New York', 'Los Angeles', 'Madison', 'Boston')) { city <- subset(cities, cityname == city_i)$city data <- readCity(city) write.table(data, file.path('data', paste(city_i, '.csv',sep='')), row.names = F, sep = ',') }
4.1.2 Pooled Dataset
################################################################ # name:Pooled Dataset setwd("~/projects/spatiotemporal-regression-models/NMMAPS-example") flist <- dir("data", full.names=T) flist <- flist[which(basename(flist) %in% c("Baton Rouge.csv", "Los Angeles.csv", "Tucson.csv", "Denver.csv") ) ] flist for(f_i in 1:length(flist)) { #f_i <- 2 fi <- flist[f_i] df <- read.csv(fi) df <- df[,c("city","date", "agecat", "cvd", "resp", "tmax", "tmin", "dptp")] # str(df) write.table(df, "outcome.csv", sep = ",", row.names = F, append = f_i > 1, col.names = f_i ==1 ) }
4.2 Exposure Data
4.3 Zones
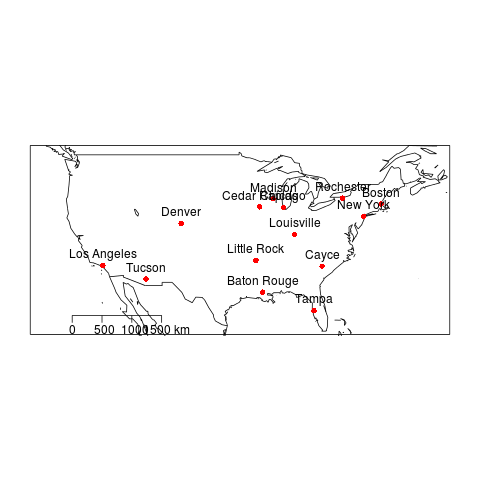
4.3.1 Map Shapefile Code
################################################################ # name:zones # func setwd("~/Dropbox/projects/spatiotemporal-regression-models/NMMAPS-example") #require(devtools) #install_github("gisviz", "ivanhanigan") require(gisviz) # load flist <- dir("data") flist # do ## geocode flist <- gsub(".csv", "", flist) flist_geo <- gGeoCode2(flist) flist_geo ## plot ## png("images/nmmaps-eg-cities.png") ## plotMyMap( ## flist_geo[,c("long","lat")], ## xl = c(-130,-60), yl = c(25,50) ## ) ## text(flist_geo$long, flist_geo$lat, flist_geo$address, pos=3) ## dev.off() # load city names flist2 <- dir("data") flist2 city_codes <- matrix(NA, nrow = 0, ncol = 2) for(fi in 1:length(flist2)) { # fi <- 1 fname <- flist2[fi] print(fi); print(fname); df <- read.csv( file.path("data", fname), stringsAsFactors = F, nrow = 1) city_codes <- rbind(city_codes, c(gsub(".csv","",fname), df$city) ) } city_codes <- as.data.frame(city_codes) names(city_codes) <- c("address","city") city_codes flist_geo2 <- merge(flist_geo, city_codes, by = "address") flist_geo2 ## make shapefile epsg <- make_EPSG() prj_code <- epsg[grep("WGS 84$", epsg$note),] prj_code shp <- SpatialPointsDataFrame(cbind(flist_geo2$long,flist_geo2$lat),flist_geo2, proj4string = CRS( epsg$prj4[which(epsg$code ==prj_code$code)] ) ) writeOGR(shp, 'cities.shp', 'cities', driver='ESRI Shapefile')
4.3.2 Map Shapefile 2
4.3.3 Map Shapefile Output
4.4 Population Data
4.4.1 Population data
- A Code Skeleton
The process for getting the population data from the websites is a bit of a pain, with repeated copy and paste operations. To make sure this is recorded I set up a skeleton to paste into.
- TODO better to use gsub() to remove the ","
################################################################ # name:population-data # func setwd("~/projects/spatiotemporal-regression-models") require(gisviz) # load ## citycodes shp <- readOGR("cities.shp", "cities") shp@data cityname <- "SELECTED CITY" # do # save url # xxx population_input <- read.table(textConnection( "age: population xxx "), sep = ":", header = TRUE) ## agecats ## 65to74 75p under65 population_input$agecat <- c(rep("under65", 10), rep("65to74"), rep("75p",2) ) citycode <- subset(shp@data, address == cityname, select = city) population_input$city <- rep( as.character(citycode[1,1]) , 13 ) population_input if(exists("population")) { population <- rbind(population, population_input ) } else { population <- population_input }
- Baton Rouge
################################################################ # name:population-data # first city, remove population data.frame rm(population) # func setwd("~/projects/spatiotemporal-regression-models") require(gisviz) # load ## citycodes shp <- readOGR("cities.shp", "cities") shp@data cityname <- "Baton Rouge" # do # save url # http://www.city-data.com/us-cities/The-South/Baton-Rouge-Population-Profile.html population_input <- read.table(textConnection( "age: population Population Under 5 years: 15502 Population 5 to 9 years: 15609 Population 10 to 14 years: 15248 Population 15 to 19 years: 21954 Population 20 to 24 years: 27230 Population 25 to 34 years: 31719 Population 35 to 44 years: 30343 Population 45 to 54 years: 27166 Population 55 to 59 years: 9495 Population 60 to 64 years: 7490 Population 65 to 74 years: 13312 Population 75 to 84 years: 9611 Population 85 years and over: 3139 "), sep = ":", header = TRUE) ## agecats ## 65to74 75p under65 population_input$agecat <- c(rep("under65", 10), rep("65to74"), rep("75p",2) ) citycode <- subset(shp@data, address == cityname, select = city) population_input$city <- rep( as.character(citycode[1,1]) , 13 ) population_input if(exists("population")) { population <- rbind(population, population_input ) } else { population <- population_input }
- Los Angeles
################################################################ # name:population-data # func #setwd("~/projects/spatiotemporal-regression-models") #require(gisviz) # load ## citycodes shp <- readOGR("cities.shp", "cities") shp@data cityname <- "Los Angeles" # do # save url # http://www.city-data.com/us-cities/The-West/Los-Angeles-Population-Profile.html population_input <- read.table(textConnection( "age: population Population under 5 years old: 285976 Population 5 to 9 years old: 297837 Population 10 to 14 years old: 255604 Population 15 to 19 years old: 251632 Population 20 to 24 years old: 299906 Population 25 to 34 years old: 674098 Population 35 to 44 years old: 584036 Population 45 to 54 years old: 428974 Population 55 to 59 years old: 143965 Population 60 to 64 years old: 115663 Population 65 to 74 years old: 18711 Population 75 to 84 years old: 125829 Population 85 years and over: 44189 "), sep = ":", header = TRUE) ## agecats ## 65to74 75p under65 population_input$agecat <- c(rep("under65", 10), rep("65to74"), rep("75p",2) ) citycode <- subset(shp@data, address == cityname, select = city) population_input$city <- rep( as.character(citycode[1,1]) , 13 ) population_input if(exists("population")) { population <- rbind(population, population_input ) } else { population <- population_input }
- Tucson
################################################################ # name:population-data # func #setwd("~/projects/spatiotemporal-regression-models") #require(gisviz) # load ## citycodes shp <- readOGR("cities.shp", "cities") shp@data cityname <- "Tucson" # do # save url # http://www.city-data.com/us-cities/The-West/Tucson-Population-Profile.html population_input <- read.table(textConnection( "age: population Population under 5 years old: 35201 Population 5 to 9 years old: 34189 Population 10 to 14 years old: 31939 Population 15 to 19 years old: 38170 Population 20 to 24 years old: 47428 Population 25 to 34 years old: 76394 Population 35 to 44 years old: 72289 Population 45 to 54 years old: 57608 Population 55 to 59 years old: 19597 Population 60 to 64 years old: 16056 Population 65 to 74 years old: 29117 Population 75 to 84 years old: 21394 Population 85 years and older: 7317 "), sep = ":", header = TRUE) ## agecats ## 65to74 75p under65 population_input$agecat <- c(rep("under65", 10), rep("65to74"), rep("75p",2) ) citycode <- subset(shp@data, address == cityname, select = city) population_input$city <- rep( as.character(citycode[1,1]) , 13 ) population_input if(exists("population")) { population <- rbind(population, population_input ) } else { population <- population_input }
- Denver
################################################################ # name:population-data # func #setwd("~/projects/spatiotemporal-regression-models") #require(gisviz) # load ## citycodes shp <- readOGR("cities.shp", "cities") shp@data cityname <- "Denver" # do # save url # http://www.city-data.com/us-cities/The-West/Denver-Population-Profile.html population_input <- read.table(textConnection( "age: population Population under 5 years old: 37769 Population 5 to 9 years old: 34473 Population 10 to 14 years old: 31315 Population 15 to 19 years old: 32259 Population 20 to 24 years old: 45534 Population 25 to 34 years old: 113676 Population 35 to 44 years old: 86420 Population 45 to 54 years old: 71000 Population 55 to 59 years old: 22573 Population 60 to 64 years old: 17191 Population 65 to 74 years old: 30643 Population 75 to 84 years old: 23369 Population 85 years and over: 8414 "), sep = ":", header = TRUE) ## agecats ## 65to74 75p under65 population_input$agecat <- c(rep("under65", 10), rep("65to74"), rep("75p",2) ) citycode <- subset(shp@data, address == cityname, select = city) population_input$city <- rep( as.character(citycode[1,1]) , 13 ) population_input if(exists("population")) { population <- rbind(population, population_input ) } else { population <- population_input }
- Louisville
################################################################ # name:population-data # func setwd("~/projects/spatiotemporal-regression-models") require(gisviz) # load ## citycodes shp <- readOGR("cities.shp", "cities") shp@data cityname <- "Louisville" # do # save url #http://www.city-data.com/us-cities/The-South/Louisville-Population-Profile.html ## NB error in table, 75-84 was missing. used same from baton rouge population_input <- read.table(textConnection( "age: population Population under 5 years old: 16926 Population 5 to 9 years old: 17359 Population 10 to 14 years old: 16627 Population 15 to 19 years old: 17362 Population 20 to 24 years old: 18923 Population 25 to 34 years old: 37541 Population 35 to 44 years old: 40354 Population 45 to 54 years old: 33755 Population 55 to 59 years old: 10716 Population 60 to 64 years old: 9211 Population 65 to 74 years old: 18577 Population 75 to 84 years: 9611 Population 85 years and older: 5075 "), sep = ":", header = TRUE) ## agecats ## 65to74 75p under65 population_input$agecat <- c(rep("under65", 10), rep("65to74"), rep("75p",2) ) citycode <- subset(shp@data, address == cityname, select = city) population_input$city <- rep( as.character(citycode[1,1]) , 13 ) population_input if(exists("population")) { population <- rbind(population, population_input ) } else { population <- population_input }
- Save Population Dataset
################################################################ # name:save-population write.csv(population, "population.csv", row.names = FALSE )
4.4.2 Population Summary
################################################################ # name:Population Summary # func require(plyr) # load population <- read.csv("population.csv") str(population) population <- population[,c("city", "agecat", "population")] # do population_summary <- ddply(population, .variables = c("city", "agecat"), .fun = summarise, sum(population) ) names(population_summary) <- c("city","agecat","pop") write.csv(population_summary, "population_summary.csv", row.names = FALSE )
4.5 Merge
################################################################ # name:merge # func setwd("~/projects/spatiotemporal-regression-models/NMMAPS-example") # load outcome <- read.csv("outcome.csv") str(outcome) outcome$date <- as.Date(outcome$date) population <- read.csv("population_summary.csv") str(population) population # do analyte <- merge(outcome, population, by = c("city", "agecat")) analyte <- arrange(analyte, city, date, agecat) # check subset(analyte, date == as.Date("1990-01-01")) # save write.csv(analyte, "analyte.csv", row.names = FALSE)
4.6 Data Checking
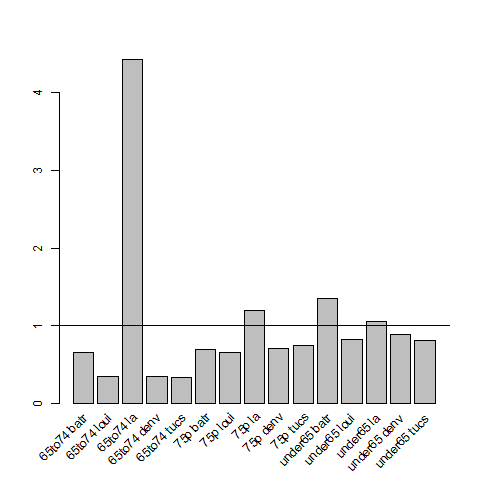
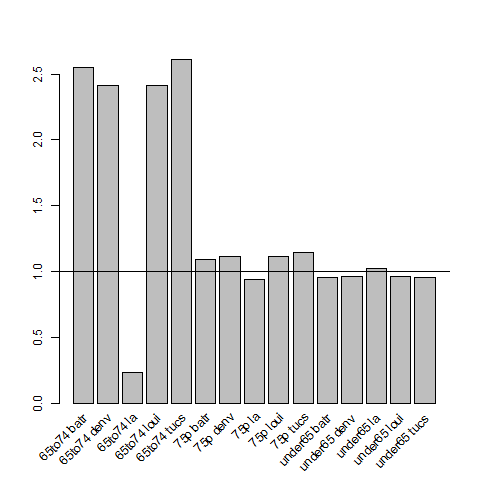
4.6.1 Check Assumption of Proportional Hazards
# Checking the proportional hazards assumption for Indirect Age Standardisation # Background ## Indirect SMRs from different index/study populations are not strictly comparable ## because they are calculated using different weighting schemes that ## depend upon the age structures of the index/study populations ## (see http://www.statsdirect.com/webhelp/#rates/smr.htm). ## Indirect SMRs can be compared if you make the assumption that the ## ratio of rates between index and reference populations is constant; ## this is similar to the assumption of proportional hazards in Cox ## regression (Armitage and Berry, 1994). ## So we need to check if the rate ratio of the study population(s) ## compared to the standard population varies substantially with age. ## If not the proportional hazards assumption holds for the standard ## rates compared with the observed rates and the Indirect SMRs are ## comparable. To do this check we calculate the Annualised Age Specific ## Rates for our study areas and for our standard for several years at ## periodic timepoints across the study period, and then calculate the ## ratio of these at each timepoint we could reassure our selves that ## this assumption holds. ## An additional issue arises when there are non-negligible differences ## in the age distributions of the study population(s) and the standard ## population. In this situation, indirect standardisation produces ## biased results due to residual confounding by age ## Also see Australian Institute of Health and Welfare. (2011). Principles on ## the use of direct age-standardisation in administrative data ## collections For measuring the gap between Indigenous and ## non-Indigenous Australians. Data linkage series. Cat. no. CSI 12. ## Canberra: AIHW. Retrieved from ## http://www.aihw.gov.au/publication-detail/?id=10737420133 ################################################################ # func setwd("~/projects/spatiotemporal-regression-models/NMMAPS-example") require(plyr) # load analyte <- read.csv("analyte.csv") # clean head(analyte) analyte$yy <- substr(analyte$date, 1, 4) # do ## first we want to see if the age specific rates vary across the ## study sites disease_colname <- "cvd" pop_colname <- "pop" by_cols <- c("city", "agecat", "yy") stdysites <- ddply(analyte, by_cols, function(df) return( c(observed = sum(df[,disease_colname]), pop = mean(df[,pop_colname]), crude.rate = sum(df[,disease_colname])/mean(df[,pop_colname]) ) ) ) ## check head(stdysites) subset(stdysites, yy == 1987 & city == "batr") ## define the subset we will use just the deaths and pop in 2000 stdy <- subset(stdysites, yy == 2000) stdy ## now define the standard population as the entire country standard <- ddply(stdy, c("agecat"), function(df) return( c(observed = sum(df[,"observed"]), pop = sum(df[,"pop"]), crude.rate = sum(df[,"observed"])/ sum(df[,"pop"]) ) ) ) standard ## Merge the studysites and the standard stdyByStnd <- merge(stdy, standard, by = "agecat") stdyByStnd ## plot the rate ratios png("images/ratio-stdy-by-stnd.png") mp <- barplot(stdyByStnd$crude.rate.x/stdyByStnd$crude.rate.y) text(mp, par("usr")[3], labels = paste(stdyByStnd$agecat,stdyByStnd$city), srt = 45, adj = c(1.1,1.1), xpd = TRUE ) abline(1,0) dev.off() ## we can see that LA has an issue with the 65to74 agecat ## Second we will check if ratio of the proportions in each population ## agecat vary between study sites and the standard totals <- ddply(stdyByStnd, c("city"), summarise, sum(pop.x) ) totals <- merge(stdyByStnd[,c("city","agecat","pop.x")], totals) totals$pop.wt <- totals[,3] / totals[,4] totals <- arrange(totals, city, agecat) totals totalsStnd <- ddply(stdyByStnd, c("agecat"), summarise, sum(pop.x) ) totalsStnd$totalPop <- sum(totalsStnd[,2]) totalsStnd$pop.wt.total <- totalsStnd[,2]/totalsStnd[,3] totalsStnd ## merge these so we can look at the ratios totals2 <- merge(totals, totalsStnd, by = "agecat") totals2 <- arrange(totals2, agecat, city) ## now plot the ratios png("images/ratio-stdy-by-stnd-pops.png") mp <- barplot(totals2$pop.wt/totals2$pop.wt.total) text(mp, par("usr")[3], labels = paste(totals2$agecat,totals2$city), srt = 45, adj = c(1.1,1.1), xpd = TRUE ) abline(1,0) dev.off() ## and we can see that there are far fewer 65to74 aged persons in LA ## than expected.
4.6.2 Incidence Rate Ratios Plot
4.6.3 Population Rate Ratios Plot
4.7 Exploratory Data Analyses
4.7.1 Time-series Plots Code
################################################################ # name:eda-tsplots # func setwd("~/projects/spatiotemporal-regression-models/NMMAPS-example") # load flist <- dir("data") fname <- flist[7]; print(fname) df <- read.csv(file.path("data", fname)) # clean str(df) summary(df$tmax); summary(df$dptp) # do ## we will only consider cities with long periods temp and humidity observed png("images/nmmaps-eg-dateranges.png", width = 1000, height=500, res = 150) par(mfrow=c(6,5), mar=c(0,3,3,0), cex=.25) for(fi in 1:length(flist)) { #fi <- 4 fname <- flist[fi] df <- read.csv(file.path("data", fname)) print(fi); print(fname); with(df, plot(as.Date(date), tmax, type = "l")) title(paste(fname, "tmax")) with(df, plot(as.Date(date), dptp, type = "l")) title(paste(fname, "dptp")) } dev.off()
4.7.2 Time-series Plots Output
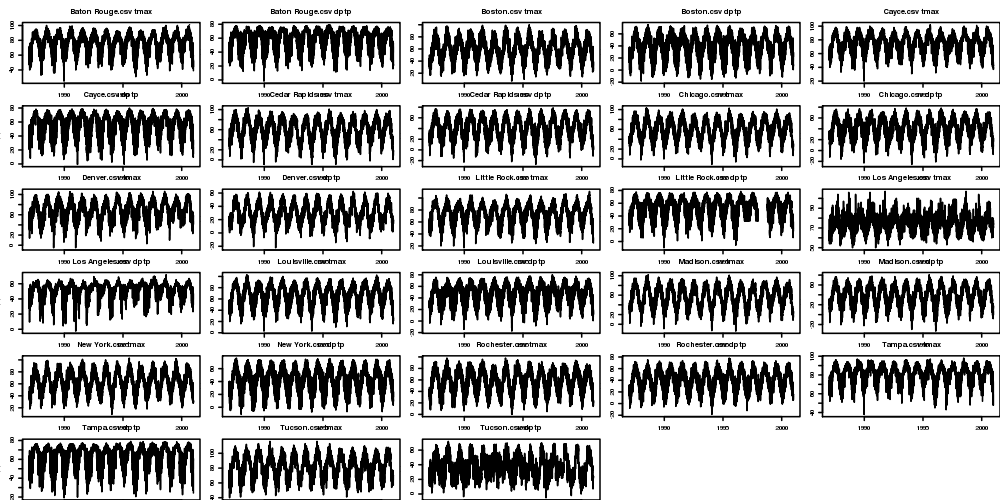
4.8 Main Analyses
4.8.1 Core Model
################################################################ # name:core # func setwd("~/projects/spatiotemporal-regression-models/NMMAPS-example") require(mgcv) require(splines) # load analyte <- read.csv("analyte.csv") # clean analyte$yy <- substr(analyte$date,1,4) numYears<-length(names(table(analyte$yy))) analyte$date <- as.Date(analyte$date) analyte$time <- as.numeric(analyte$date) analyte$agecat <- factor(analyte$agecat, levels = c("under65", "65to74", "75p"), ordered = TRUE ) # do fit <- gam(cvd ~ s(tmax) + s(dptp) + city + agecat + s(time, k= 7*numYears, fx=T) + offset(log(pop)), data = analyte, family = poisson ) # plot of response functions png("images/nmmaps-eg-core.png", width = 1000, height = 750, res = 150) par(mfrow=c(2,3)) plot(fit, all.terms = TRUE) dev.off()
4.8.2 Core Model Plots
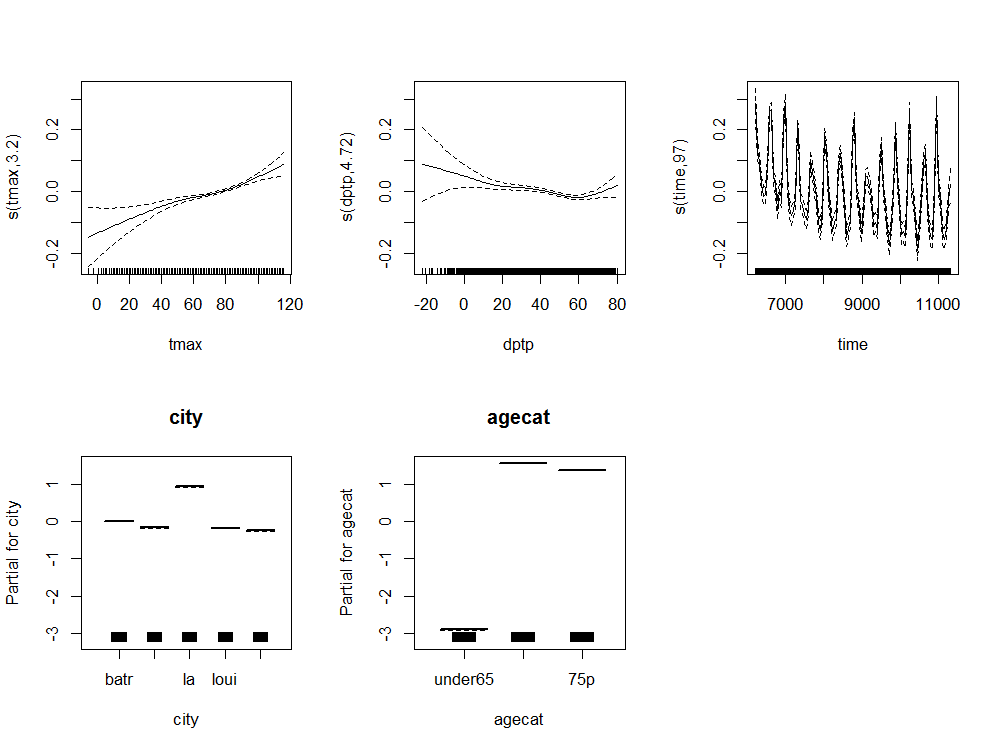
4.8.3 Model Selection
# The following codes are just a dump of stuff I've found useful in the past. # This is all time-series methods. # TODO replace the city-specific model with a pooled analysis with city specific trend. # func require(mgcv) require(splines) ###################################################### # load dir("data") city <- "Chicago" data <- read.csv(sprintf("data/%s.csv", city), header=T) str(data) data$yy <- substr(data$date,1,4) data$date <- as.Date(data$date) ###################################################### # check par(mfrow=c(2,1), mar=c(4,4,3,1)) with(subset(data[,c(1,15:25)], agecat == '75p'), plot(date, tmax) ) with(subset(data[,c(1,4,15:25)], agecat == '75p'), plot(date, cvd, type ='l', col = 'grey') ) with(subset(data[,c(1,4,15:25)], agecat == '75p'), lines(lowess(date, cvd, f = 0.015)) ) # I am worried about that outlier data$date[which(data$cvd > 100)] # [1] "1995-07-15" "1995-07-16" ###################################################### # do standard NMMAPS timeseries poisson GAM model numYears<-length(names(table(data$yy))) df <- subset(data, agecat == '75p') df$time <- as.numeric(df$date) fit <- gam(cvd ~ s(pm10tmean) + s(tmax) + s(dptp) + s(time, k= 7*numYears, fx=T), data = df, family = poisson) # plot of response functions par(mfrow=c(2,2)) plot(fit) dev.off() ###################################################### # some diagnostics summary(fit) # note the R-sq.(adj) = 0.21 gam.check(fit) # note the lack of a leverage plot. for that we need glm ###################################################### # do same model as glm fit2 <- glm(cvd ~ pm10tmean + ns(tmax, df = 8) + ns(dptp, df = 4) + ns(time, df = 7*numYears), data = df, family = poisson) # plot responses par(mfrow=c(2,2)) termplot(fit2, se =T) dev.off() # plot prediction df$predictedCvd <- predict(fit2, df, 'response') # baseline is given by the intercept fit3 <- glm(cvd ~ 1, data = df, family = poisson) df$baseline <- predict(fit3, df, 'response') with(subset(df, date>=as.Date('1995-01-01') & date <= as.Date('1995-07-31')), plot(date, cvd, type ='l', col = 'grey') ) with(subset(df, date>=as.Date('1995-01-01') & date <= as.Date('1995-07-31')), lines(date,predictedCvd) ) with(subset(df, date>=as.Date('1995-01-01') & date <= as.Date('1995-07-31')), lines(date,baseline) ) ###################################################### # some diagnostics # need to load a function to calculate poisson adjusted R squared # original S code from # The formula for pseudo-R^2 is taken from G. S. Maddalla, # Limited-dependent and Qualitative Variables in Econometrics, Cambridge:Cambridge Univ. Press, 1983. page 40, equation 2.50. RsquaredGlm <- function(o) { n <- length(o$residuals) ll <- logLik(o)[1] ll_0 <- logLik(update(o,~1))[1] R2 <- (1 - exp(((-2*ll) - (-2*ll_0))/n))/(1 - exp( - (-2*ll_0)/n)) names(R2) <- 'pseudo.Rsquared' R2 } RsquaredGlm(fit2) # 0.51 # the difference is presumably due to the arguments about how to account for unexplainable variance in the poisson distribution? # significance of spline terms drop1(fit2, test='Chisq') # also note AIC. best model includes all of these terms # BIC can be computed instead (but still labelled AIC) using drop1(fit2, test='Chisq', k = log(nrow(data))) # diagnostic plots par(mfrow=c(2,2)) plot(fit2) dev.off() # note high leverage plus residuals points are labelled # leverage doesn't seem to be too high though which is good # NB the numbers refer to the row.names attribute which still refer to the original dataset, not this subset df[row.names(df) %in% c(9354,9356),]$date # as suspected [1] "1995-07-15" "1995-07-16" ###################################################### # so lets re run without these obs df2 <- df[!row.names(df) %in% c(9354,9356),] # to avoid duplicating code just re run fit2, replacing data=df with df2 # tmax still significant but not so extreme # check diagnostic plots again par(mfrow=c(2,2)) plot(fit2) dev.off() # looks like a well behaved model now. # if we were still worried about any high leverage values we could identify these with df3 <- na.omit(df2[,c('cvd','pm10tmean','tmax','dptp','time')]) df3$hatvalue <- hatvalues(fit2) df3$res <- residuals(fit2, 'pearson') with(df3, plot(hatvalue, res)) # this is the same as the fourth default glm diagnostic plot, which they label x-axis as leverage summary(df3$hatvalue) # gives us an idea of the distribution of hat values # decide on a threshold and look at it hatThreshold <- 0.1 with(subset(df3, hatvalue > hatThreshold), points(hatvalue, res, col = 'red', pch = 16)) abline(0,0) segments(hatThreshold,-2,hatThreshold,15) dev.off() fit3 <- glm(cvd ~ pm10tmean + ns(tmax, df = 8) + ns(dptp, df = 4) + ns(time, df = 7*numYears), data = subset(df3, hatvalue < hatThreshold), family = poisson) par(mfrow=c(2,2)) termplot(fit3, se = T) # same same plot(fit3) # no better # or we could go nuts with a whole number of ways of estimating influence # check all influential observations infl <- influence.measures(fit2) # which observations 'are' influential inflk <- which(apply(infl$is.inf, 1, any)) length(inflk) ###################################################### # now what about serial autocorrelation in the residuals? par(mfrow = c(2,1)) with(df3, acf(res)) with(df3, pacf(res)) dev.off() ###################################################### # just check for overdispersion fit <- gam(cvd ~ s(pm10tmean) + s(tmax) + s(dptp) + s(time, k= 7*numYears, fx=T), data = df, family = quasipoisson) summary(fit) # note the Scale est. = 1.1627 # alternatively check the glm fit2 <- glm(cvd ~ pm10tmean + ns(tmax, df = 8) + ns(dptp, df = 4) + ns(time, df = 7*numYears), data = df, family = quasipoisson) summary(fit2) # (Dispersion parameter for quasipoisson family taken to be 1.222640) # this is probably near enough to support a standard poisson model... # if we have overdispersion we can use QAIC (A quasi- mode does not have a likelihood and so does not have an AIC, by definition) # we can use the poisson model and calculate the overdispersion fit2 <- glm(cvd ~ pm10tmean + ns(tmax, df = 8) + ns(dptp, df = 4) + ns(time, df = 7*numYears), data = df, family = poisson) 1- pchisq(deviance(fit2), df.residual(fit2)) # QAIC, c is the variance inflation factor, the ratio of the residual deviance of the global (most complicated) model to the residual degrees of freedom c=deviance(fit2)/df.residual(fit2) QAIC.1=-2*logLik(fit2)/c + 2*(length(coef(fit2)) + 1) QAIC.1 # Actually lets use QAICc which is more conservative about parameters, QAICc.1=-2*logLik(fit2)/c + 2*(length(coef(fit2)) + 1) + 2*(length(coef(fit2)) + 1)*(length(coef(fit2)) + 1 + 1)/(nrow(na.omit(df[,c('cvd','pm10tmean','tmax','dptp','time')]))- (length(coef(fit2))+1)-1) QAICc.1 ###################################################### # the following is old work, some may be interesting # such as the use of sinusoidal wave instead of smooth function of time # # sine wave # timevar <- as.data.frame(names(table(df$date))) # index <- 1:length(names(table(df$date))) # timevar$time2 <- index / (length(index) / (length(index)/365.25)) # names(timevar) <- c('date','timevar') # timevar$date <- as.Date(timevar$date) # df <- merge(df,timevar) # fit <- gam(cvd ~ s(tmax) + s(dptp) + sin(timevar * 2 * pi) + cos(timevar * 2 * pi) + ns(time, df = numYears), data = df, family = poisson) # summary(fit) # par(mfrow=c(3,2)) # plot(fit, all.terms = T) # dev.off() # # now just explore the season fit # fit <- gam(cvd ~ sin(timevar * 2 * pi) + cos(timevar * 2 * pi) + ns(time, df = numYears), data = df, family = poisson) # yhat <- predict(fit) # head(yhat) # with(df, plot(date,cvd,type = 'l',col='grey', ylim = c(15,55))) # lines(df[,'date'],exp(yhat),col='red') # # drop1(fit, test= 'Chisq') # # drop1 only works in glm? # # fit with weather variables, use degrees of freedom estimated by gam # fit <- glm(cvd ~ ns(tmax,8) + ns(dptp,2) + sin(timevar * 2 * pi) + cos(timevar * 2 * pi) + ns(time, df = numYears), data = df, family = poisson) # drop1(fit, test= 'Chisq') # # use plot.glm for diagnostics # par(mfrow=c(2,2)) # plot(fit) # par(mfrow=c(3,2)) # termplot(fit, se=T) # dev.off() # # cyclic spline, overlay on prior sinusoidal # with(df, plot(date,cvd,type = 'l',col='grey', ylim = c(0,55))) # lines(df[,'date'],exp(yhat),col='red') # df$daynum <- as.numeric(format(df$date, "%j")) # df[360:370,c('date','daynum')] # fit <- gam(cvd ~ s(daynum, k=3, fx=T, bs = 'cp') + s(time, k = numYears, fx = T), data = df, family = poisson) # yhat2 <- predict(fit) # head(yhat2) # lines(df[,'date'],exp(yhat2),col='blue') # par(mfrow=c(1,2)) # plot(fit) # # fit weather with season # fit <- gam(cvd ~ s(tmax) + s(dptp) + # s(daynum, k=3, fx=T, bs = 'cp') + s(time, k = numYears, fx = T), data = df, family = poisson) # par(mfrow=c(2,2)) # plot(fit) # summary(fit)
5 Spatial lag and timeseries
I will use the same NMMAPSlite to show how I'd approach a simple "Spatio-Temporal" model.
- TODO The following is a stub of an idea. For further development
5.1 Data
5.1.1 Zones
- Spatial Neighbours Code
################################################################ # name:spatwat # func setwd("~/projects/spatiotemporal-regression-models/NMMAPS-example") require(gisviz) # load dir() shp <- readOGR("cities.shp", "cities") # clean head(shp@data) # do ## I will use nearest neighbour within a distance threshold, but ## usually polygon datasets would use poly2nb nb <- dnearneigh(shp, d1 = 1, d2 = 1000) head(nb) shp[[1]][1] shp[[1]][nb[[1]]] # map nudge <- 10 png("images/nmmaps-eg-neighbourhood.png") plotMyMap( shp@coords, xl = c(min(shp@coords[,1])-nudge, max(shp@coords[,1])+nudge), yl = c(min(shp@coords[,2])-nudge, max(shp@coords[,2])+nudge) ) plot(nb, shp@coords, add=TRUE) text(shp@data$long, shp@data$lat, shp@data$address, pos=3) points(shp@coords[nb[[1]],], col = 'green', pch = 16) points(shp@data[1,c("long","lat")],col = 'blue', pch = 16) dev.off()
- Spatial Neighbours Output
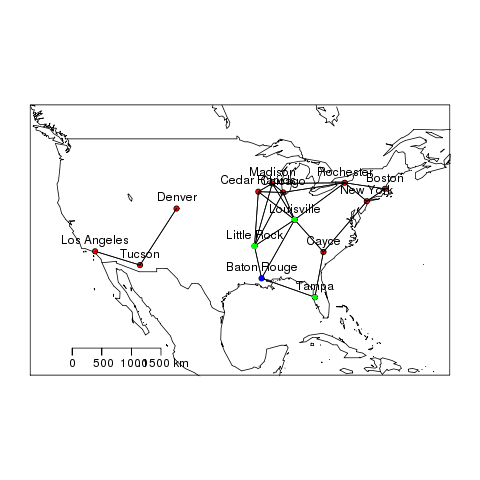
5.1.2 Outcome
#### Now do the Neighbourhood autoregressive lagged variable #### # func setwd("~/projects/spatiotemporal-regression-models/NMMAPS-example") require(gisviz) require(plyr) # load analyte <- read.csv("analyte.csv") shp <- readOGR("cities.shp", "cities") # clean analyte$yy <- substr(analyte$date,1,4) numYears<-length(names(table(analyte$yy))) analyte$date <- as.Date(analyte$date) analyte$time <- as.numeric(analyte$date) analyte$agecat <- factor(analyte$agecat, levels = c("under65", "65to74", "75p"), ordered = TRUE ) names(analyte) analyte[1,] table(analyte$city) # do study <- analyte[, c("city","date", "agecat", "cvd", "pop")] subset(study, city == "tucs" & date == as.Date("1991-01-01")) nb <- dnearneigh(shp, d1 = 1, d2 = 1000) head(nb) head(shp@data) adj <- adjacency_df(NB = nb, shp = shp, zone_id = 'city') subset(adj, V1 == "tucs") neighbours <- merge(study, adj, by.x = "city", by.y = "V1") subset(neighbours, city == "tucs" & date == as.Date("1991-01-01")) xvars <- c("V2", "date","agecat") yvars <- c("city", "date", "agecat") neighbours <- merge(neighbours[,c(xvars, "city")], analyte[,c(yvars, "cvd", "pop")], by.x = xvars, by.y = yvars) names(neighbours) subset(neighbours, city == "tucs" & date == as.Date("1991-01-01")) neighbours$asr <- (neighbours$cvd / neighbours$pop) * 1000 neighbours2 <- ddply(neighbours, c("city", "date", "agecat"), summarise, NeighboursYij = mean(asr) ) subset(neighbours2, city == "tucs" & date == as.Date("1991-01-01")) table(neighbours2$city) head(analyte) analyte <- merge(analyte, neighbours2, by = c("city", "agecat", "date")) analyte <- arrange(analyte, city, date, agecat) head(analyte)
5.2 Analysis
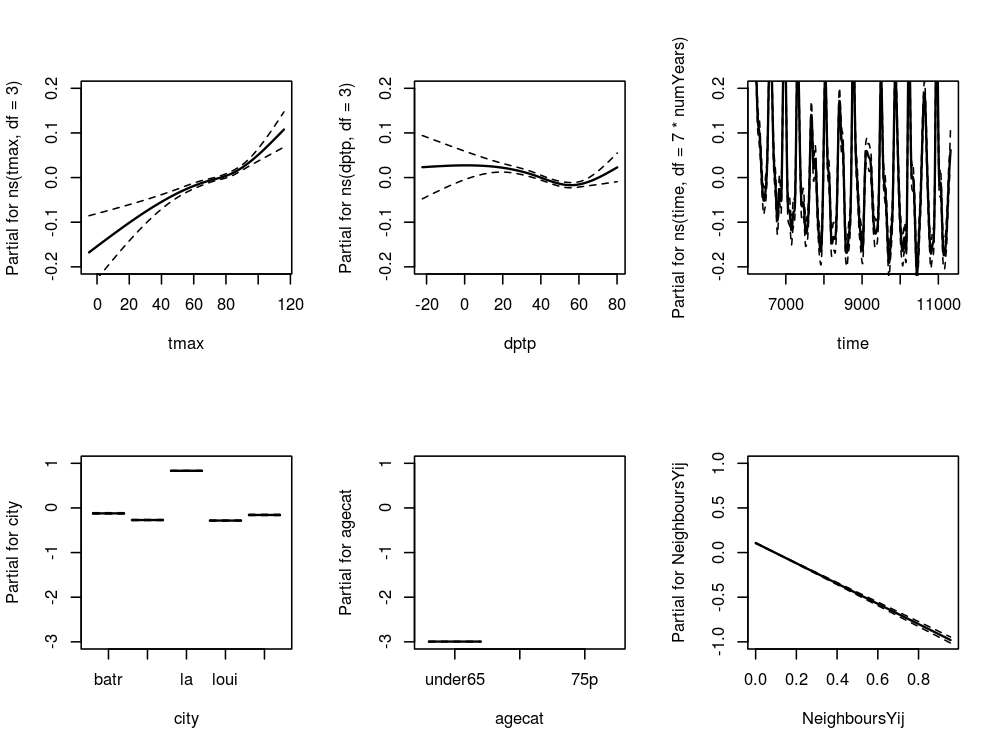
5.2.1 Model with Spatial Lag
# func require(splines) # load # assumes the prior code chunks have been run # do fit <- glm(cvd ~ ns(tmax, df = 3) + ns(dptp, df = 3) + NeighboursYij + city + agecat + ns(time, df = 7*numYears) + offset(log(pop)), data = analyte, family = poisson ) # plot of response functions png("images/nmmaps-eg-sp-lag.png", width = 1000, height = 750, res = 150) par(mfrow=c(2,3)) termplot(fit, terms = attr(terms(fit),'term.labels') [c(1:2,6)], se = TRUE, ylim =c(-.2,.2), col.term = 'black', col.se = 'black') termplot(fit, terms = attr(terms(fit),'term.labels') [c(4,5)], se = TRUE, ylim =c(-3,3), col.term = 'black', col.se = 'black') termplot(fit, terms = attr(terms(fit),'term.labels') [3], se = TRUE, ylim =c(-1,1), col.term = 'black', col.se = 'black') dev.off()
5.2.2 Spatial Lag Model Plots